A.A. Al-Mahbashi,1 M.M. Mukhtar 2 and E.S. Mahgoub 3
تحديد الأنماط الجزيئية للمستفرَدات من أنواع المتفطِّرات لدى مرضى السل في اليمن
أنس أحمد المحبشي، معاوية مختار، الشيخ محجوب
الخلاصـة: أجرى الباحثون هذه الدراسة للتعرُّف على أنواع المتفطِّرات المستفرَدة من مرضى السل الرئوي في اليمن، وقد جمع الباحثون عينات البلغم في الصباح الباكر من 170 مريضاً تم تحويلهم إلى المعهد الوطني للسل في صنعاء للاشتباه بإصابتهم بالسل الرئوي، وقد تم تلوين العينات بملوّن تسيل نيلسون وزرعت في مستنبَط أوغاوا ولفينشتين – جنسن، وتم تضخيم المتتاليات المستهدفة في الجين rpoB باستخدام مواد تسبب الطفرة للبادئات للأمام وللخلف، ثم تلا ذلك استخدام الإنزيم HindIII للهضم. ومن بين 120 مستفرَدة حللها الباحثون تبين أن 118 منها (%98.3) قد تم التعرُّف عليها على أنها معقَّد المتفطرة السلية، وأن 2 منها (%1.7) تم التعرُّف عليهما على أنهما متفطرتان غير سليتين. وتوضح النتائج التي توصل إليها الباحثون أن هاتين المستفرَدتين هما من المتفطرات المقاومة للأدوية، كما أن تحليل متتاليات الدنا أظهر أن تراصف الحمض النووي في الدنا في المستفرَدات من المتفطرات غير السلية كان يختلف عما هو عليه في معقد المتفطرات السلية.
ABSTRACT This study was done to characterize at the species level Mycobacterium spp. isolates from Yemeni pulmonary tuberculosis patients. Early-morning sputum samples were collected from 170 patients referred to the National Tuberculosis Institute in Sana’a city with suspected pulmonary tuberculosis. Samples were processed with Ziehl–Neelsen stain and cultured in Ogawa and Lowenstein–Jensen media. The rpoB gene target sequence was amplified using mutagenesis forward and reverse primers followed by HindIII enzyme digestion. Of the 120 isolates analysed, 118 (98.3%) were identified as M. tuberculosis complex and 2 (1.7%) were identified as mycobacteria other than M. tuberculosis. The results showed that those 2 isolates were multi-drug resistant and the DNA sequencing analysis showed that the alignment of nucleic acid of DNA in isolates of mycobacteria other than M. tuberculosis was different from that of M. tuberculosis complex.
Typage moléculaire des isolats de Mycobacterium spp. prélevés chez des patients yéménites atteints de tuberculose
RÉSUMÉ La présente étude a été menée afin de caractériser l’espèce des isolats de Mycobacterium spp. prélevés chez des patients yéménites atteints de tuberculose. Des échantillons d’expectoration ont été prélevés tôt le matin chez 170 patients qui avaient été orientés vers l’Institut national de la tuberculose de la ville de Sanaa pour suspicion de tuberculose pulmonaire. Les échantillons ont été traités par coloration de Ziehl-Neelsen et mis en culture sur milieux Löwenstein–Jensen et Ogawa. La séquence cible du gène rpoB a été amplifiée selon la méthode des amorces mutagènes directe et inverse suivie par une digestion par l’enzyme HindIII. Sur les 120 isolats analysés, 118 (98,3 %) ont été identifiés comme appartenant au complexe M. tuberculosis et 2 (1,7 %) comme étant des mycobactéries d’un autre type que M. tuberculosis. Nos résultats ont révélé que ces deux isolats étaient pharmacorésistants tandis que l’analyse des séquences d’ADN a montré que l’alignement d’acide nucléique dans les isolats des mycobactéries d’un autre type que M. tuberculosis était différent de celui du complexe M. tuberculosis.
1Department of Microbiology, Faculty of Science, University of Sana’a, Sana’a, Yemen.
2Institute of Endemic Disease;
3Department of Microbiology and Parasitology, Faculty of Medicine, University of Khartoum, Khartoum, Sudan (Correspondence to E.S. Mahgoub:
Received: 04/06/12; accepted: 25/09/12
EMHJ, 2013, 19(11):942-946
Introduction
Tuberculosis (TB) is a disease of major public health concern worldwide. It is a bacterial infectious disease that is considered the second most important cause of death due to an identifiable infectious agent [1]. Approximately one-third of the world’s population is infected with latent TB and 5%–10% of this population will develop active stages of the disease during their life time [2].
TB is a highly transmissible disease and infection can occur via inhalation of droplet particles aerosolized from persons infected with Mycobacterium tuberculosis or by consumption of milk infected with bovine M. bovis. The disease can infect humans and animals, with outcomes ranging from localized lesions to disseminated disease. The genus Mycobacterium comprises more than 70 species, some of which are potentially pathogenic to humans and animals and some of which are saprophytic. Mycobacteria that cause TB in mammals form the Mycobacterium tuberculosis complex (MTC) and include M. tuberculosis, M. africanum, M. bovis or M. bovis BCG, M. microtti and M. canetti. Other forms of mycobacteria that are considered opportunistic are termed mycobacteria other than Mycobacterium tuberculosis (MOTT) [3].
Yemen is one of the poorest of the world’s low-income countries and TB is one of the most infectious diseases that are endemic in the Yemeni population. The absolute number of TB cases in Yemen is not known, but 37 000 cases were recorded as under treatment throughout the country in the year 2002 [4]. The main objective of this study was to use molecular techniques to identify and characterize Mycobacterium spp. isolated from pulmonary TB patients in Yemen.
Methods
Sample collection
The study was conducted on patients referred to the National Tuberculosis Institute in Sana’a city with suspected pulmonary TB based on their presentation with cough more than 2 weeks. The Institute is a specialist referral centre for TB diagnosis and therapy and is situated in Sana’a the capital city of Yemen. The Institute receives TB patients from all regions of Yemen and provides free treatment.
An early-morning sputum sample was collected from 170 patients into wide-mouthed plastic containers. Baseline data of the patients was collected by completion of a questionnaire administered during the collection of samples. Samples were collected between January 2004 and October 2005 and the study was completed in 2008.
The study was approved by the University of Sana’a ethics committee and consent was obtained from the participants before their enrolment in the study.
Laboratory methods
Antibiotic sensitivity testing, using standard methods, was carried out on cultures from all 170 samples. For cost reasons, PCR was done on only 120 of the samples.
Staining and culture methods
Sputum samples were treated with 4% NaOH and stained by Ziehl–Neelsen stain to detect acid-fast bacilli [5]. The sputum samples were cultured on a special egg-based solid medium (Ogawa medium) according to the procedures of the Japan International Cooperation Agency [6]. Typically growth of Mycobacteria spp. appears within 3–4 weeks. The colonies are buff in colour with a dry and friable surface and irregular edges.
DNA extraction
Two colonies were taken from the culture medium and placed in 100 µL of sterile distilled water; 100 µL of phenol-chloroform reagent was added and the mixture was vortexed for about 10 s and heated at 80 °C for 20 min. The mixture was stored at –20 °C in microcentrifuge tubes (free from DNA or RNA) until needed [7].
Polymerase chain reaction technique
Primers specific for the rpoB gene, encoding the B-subunit of RNA polymerase (rpoB DNA, 342–360 base pairs) was the target region for amplification and identification of Mycobacterium spp. [8]. The polymerase chain reaction (PCR) mixture was prepared as follows: distilled water 5.0 µL, sample DNA 4.0 µL, PCR buffer 2.5 µL, PCR MgCl2 2.0 µL, PCR dNTP 2.5 µL, primers 3 µL, Tag polymerase 1.0 µL. The PCR mixture was gently mixed and amplified using a thermocycler (Perkin Elmer) adjusted to the cycling programme for 30 cycles. The sequence of rpoB primers for the mutagenesis forward primer was
5′-CGA CCA CTT CGG CAA CCG-3′ and for the mutagenesis reverse primer was 5′-TCG ATC GGG CAC ATC CGG-3′.
Restriction fragment length polymorphism (RFLP)
Following amplification of the rpoB gene the product was subjected to digestion by HindIII restriction enzymes (Roche) as follows: 15 µL from PCR product was pipetted into PCR tubes, 2 µL of enzyme was added to the tube, 2 µL of enzyme buffer was also added to the tube, then 1 µL of distilled water was added to the mixture and mixed well [9].
DNA sequencing analysis
PCR-amplified DNA of the drug-resistant isolates was commercially sequenced by Macrogen Company using the BigDye terminator cycling and universal primers.
Results
The age of the whole sample of 170 patients ranged between 12–70 years old, and the largest proportion was in the age group 20–30 years (Table 1). There were significantly more males (117, 69%) than females (53, 31%) (P < 0.05).
Our results showed that mycobacterial DNA was amplified successfully using the relevant primers and the size of DNA was 360 bp compared with the molecular weight marker (100 bp) (Figure 1).
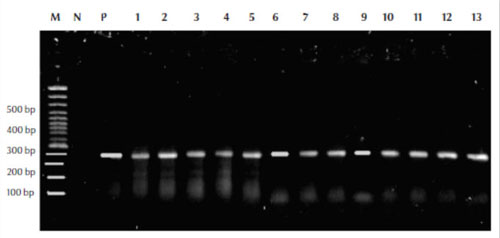
Figure 1 Polymerase chain reaction assay results using the MF and MR primers for detection and amplification of the Mycobacterium rpoB gene from the isolates with leader marker (100 bp) to detect the size of amplified DNA M = molecular weight control marker (100 bp); P = positive control; N = negative control; lanes 1–13 from patient samples
The PCR-RFLP results showed that 118/120 (98.3%) of the isolates were MTC, whereas 2/120 (1.7%) were MOTT (Figure 2A and B).
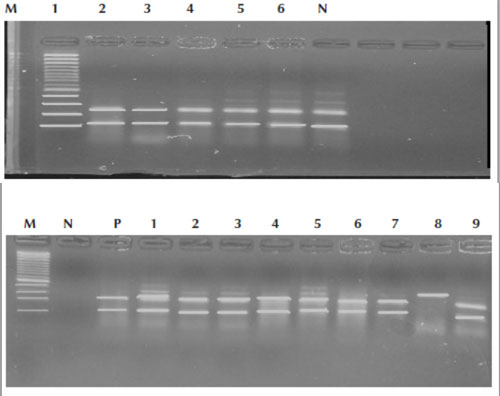
Figure 2A Polymerase chain reaction–restriction fragment length polymorphism assay using restriction enzymes of confirmed tuberculosis patients; lanes 1,2,3,4,5,6 were Mycobacterium tuberculosis complex isolates that showed 2 fragments. M = control marker 100 bp; N = negative control
Figure 2B Polymerase chain reaction–restriction fragment length polymorphism assay using restriction enzymes of confirmed tuberculosis patients; lanes 1,2,3,4,5,6,7,9 were Mycobacterium tuberculosis complex isolates that showed 2 fragments, whereas lane 8 is Mycobacteria other than M. tuberculosis that showed 1 fragment. M = control marker 100 bp; N = negative control; P = positive control
The DNA sequencing analysis results showed that the DNA sequence of MTC strains were different from MOTT strains (Figure 3).

Figure 3 DNA sequence analysis alignment showing the difference between nucleic acid of Mycobacterium tuberculosis complex (samples 1–12) and Mycobacteria other than M. tuberculosis (sample 13)
Discussion
A recent health report on Arab countries by the World Health Organization declared that TB was an important public health problem in the 19 Arab countries of the Eastern Mediterranean Region, affecting 240 000 people with 53 000 deaths every year; 85% of the deaths occurred in adults [10]. In Yemen, TB is considered one of the major infectious diseases recorded in the national disease list [11]. A rapidly increasing population, poor quality health services, very low annual income of individuals and the whole country’s poor economic status are the most important factors responsible for the high incidence of TB in the country [12].
In the present study 170 patients were recruited who were suspected of having pulmonary TB based on their presentation with cough more than 2 weeks. The age of the patients ranged between 12–70 years old, and the highest prevalence was among the age group 20–30 years. These results are in agreement with previous reports from the national disease surveillance infectious diseases centre and the Ministry of Health [11,12]. As previously reported, in this study males were significantly more affected than females [13]. In Yemen the higher rate of TB in males may be attributed to the different habits of males, especially smoking the waterpipe (nargile or mada’a), which is usually shared between different persons. Another prevalence study on pulmonary TB attributed the low prevalence of TB among women to underdetection of TB in females because women often choose medical care providers operating outside the national TB control centres [14].
Routine sputum smears and Mycobacterium spp. cultures confirmed the presence of acid-fast bacilli in the 170 sputum samples examined. However, acid-fast staining does not identify the Mycobacterium to the species level. In addition the time required to detect the organism by routine culture is approximately 4–8 weeks. PCR assay was therefore used to characterize 120 of the isolates. The rpoB gene was successfully amplified in all isolates and enabled identification of Mycobacterium spp. following restriction of the PCR product by HindIII restriction enzyme that produced 2 DNA fragments in the amplicons of M. tuberculosis.
The PCR-RFLP results identified 98.3% of isolates as MTC and 1.7% samples as MOTT. Interestingly the MOTT isolates were resistant to isoniazid, rifampicin and streptomycin. MOTT have been reported to cause infection in humans and to complicate treatment regimens since they may not respond to routinely used anti-TB drugs [15–17]. Based on the results of this study we recommend the use of molecular techniques for identification of the Mycobacterium spp. before initiation of treatment.
Acknowledgements
Funding: This research was supported by the Ministry of Higher Education of Yemen as part of a PhD degree for the first author.
Competing interests: None declared.
References
- Tiruviluamala P, Reichman LB. Tuberculosis. Annual Review of Public Health, 2002, 23:403–426.
- Jones-Lopez EC, Ellner JJ. Tuberculosis and atypical mycobacterial infections. In: Guerrant RL, Walker DH, Weller PF, eds. Tropical infectious diseases: principles pathogens and practice. New York, Elsevier, 2011 (Chapter 36).
- Annual heath report. National tuberculosis control programme. Sana’a, Yemen, Ministry of Health, 2004.
- Edsel M, Gregory SC, Robert AS. Mycobacterium other than tuberculosis (MOTT) infection: an emergency disease in infliximab treated patients. Journal of Infection, 2007, 10:1–4.
- Cheesbrough M. District laboratory practice in tropical countries. Volume 2. Cambridge, Cambridge University Press, 2000:207–213.
- Kawai M, Fujiki A. Minimum essentials of laboratory procedure for tuberculosis control. Tokyo, Japan Anti-Tuberculosis Association, Research Institute of Tuberculosis in Japan, 1996.
- Yates MD, Drobniewski FA, Wilson.SM. Evaluation of a rapid PCR-based epidemiological typing method for routine studies of Mycobacterium tuberculosis. Journal of Clinical Microbiology, 2002, 40(2):712–714.
- Kim BJ et al. Differentiation of mycobacterial species by PCR-restriction analysis of DNA (342 base pairs) of the RNA polymerase gene (rpoB). Journal of Clinical Microbiology, 2001, 39:2102–2109.
- Kim BJ et al. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB). Journal of Clinical Microbiology, 1999, 37:1714–1720.
- Tuberculosis. In: Overview of child health in Arab countries, 2nd ed. Alexandria, Egypt, World Health Organization Regional Office for the Eastern Mediterranean, 2002.
- Tuberculosis infection: disease surveillance of infectious disease. Sana’a, Yemen, Ministry of Health and World Health Organization Country Office, 2000:49–52.
- National tuberculosis programme. Tuberculosis in Republic of Yemen. Sana’a, Yemen, Ministry of Health, 1996.
- Abassi A, Mansourian AR. Efficacy of DOTS strategies in treatment of respiratory tuberculosis in Gorgan, Islamic Republic of Iran. Eastern Mediterranean Health Journal, 2007, 13(3):664–669.
- Thorson A et al. Do women with tuberculosis have a lower likelihood of getting diagnosed? Prevalence and case detection of sputum smear positive pulmonary TB, a population-based study from Vietnam. Journal of Clinical Epidemiology, 2004, 57(4):398–402.
- Kearns AM et al. Epidemiology and molecular typing of an outbreak of tuberculosis in a hostel for homeless men. Journal of Clinical Pathology, 2000, 53(2):122–124.
- Sharaf-Eldin GS et al. Molecular analysis of clinical isolates of Mycobacterium tuberculosis collected from patients with persistent disease in the Khartoum region of Sudan. Journal of Infection, 2002, 44:244–251.
- Sanguinetti M et al. Routine use of PCR-reverse cross-blot hybridization assay for rapid identification of Mycobacterium species growing in liquid media. Journal of Clinical Microbiology, 1998, 36:1530–1533.


