Research article
H. Hajjaran,1 B. Azarian,2 M. Mohebali,1 R. Hadighi,3 A. Assareh1 and B. Vaziri2
دراسة بروتيوميّة مقارنة حول الليشمانيات المدارية الحساسة لأنتيمونات الميغلومين وتلك المستعصية عليها من المستفرَدة مرضى تلك من الإيرانيين المرضى بداء الليشمانيات الجلدية البشرية
هما حجاران، بهاره آذريان، مهدي محبعلي، رامتين حديقي، آرزو عصاره، بهروز وزيري
الخلاصة: في سبيل تحديد تغيُّرات التعبير عن الروتينات، المتعلّقة باستعصاء الليشمانيات الجلدية على أنتيمونات الميغلومين، فقد أجرى الباحثون تحاليل بروتيومية مقارنة للذّراري المستعصية والذّراري الحساسة من الليشمانيات المدارية المستفرَدة من مرضى إيرانيين، باستخدام الرحلان الكهربي الثنائي الأبعاد للبروتينات الخلوية، ومن ثَمَّ تحديد البروتينات التي يُعَبَّر عنها تعبيراً مختلفاً باستخدام القياس الطيفي الكُتْلَويّ لزمن الارتشاف والتأيُّن بالليزر الـمُساعَد بالمصفوفة. وقد أدى تحليل الصور للخرائط الموافقة إلى التعرف على سبعة بروتينات مفرطة التعبير أو ناقصة التعبير؛ وهذه البروتينات هي: مستقبل بروتين الكيناز سي المفعَّل، والألفا توبولين، ومُخَلِّقة البروستا غلاندين إف 2 ألفا، ومصاوِغَة ثنائي سلفيد البروتين، وبروتين النقل الحويصلي، بالإضافة إلى بروتين افتراضي. وقد أظهرت الدراسة مدى فائدة دراسة البروتيوميات في التعرف على البروتينات التي يمكن أن تعبِّر عن الفوارق بين المستفرَدات الحساسة والمستفرَدات المستعصية من الليشمانية المدارية.
ABSTRACT In order to define the protein expressional changes related to the process of meglumine antimoniate resistance in anthroponotic cutaneous leishmaniasis (CL), we performed a comparative proteomics analysis on sensitive and resistant strains of Leishmania tropica isolated from Iranian CL patients. Cell proteins were analysed with 2-dimensional electrophoresis and differentially expressed proteins were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Image analysis of the matched maps identified 7 proteins that were either over- or down-expressed: activated protein kinase c receptor (LACK), alpha tubulin (×2), prostaglandin f2-alpha synthase, protein disulfide isomerase, vesicular transport protein and a hypothetical protein. The study shows the usefulness of proteomics in identifying proteins that may express differences between sensitive and resistant L. tropica isolates.
étude protéomique comparative de souches Leishmania tropica sensibles ou résistantes à l’antimoniate de méglumine chez des patients iraniens atteints de leishmaniose cutanée anthroponotique
RÉSUMÉ Afin de déterminer les modifications de l’expression des protéines liées au processus de résistance à l’antimoniate de méglumine dans la leishmaniose cutanée anthroponotique, nous avons réalisé une analyse protéomique comparative de souches sensibles et de souches résistantes de Leishmania tropica isolées à partir d’échantillons prélevés chez des patients iraniens atteints de la maladie. Les protéines cellulaires ont été analysées par électrophorèse bidimensionnelle ; des protéines différentiellement exprimées ont été identifiées par spectrométrie de masse à temps de vol avec désorption-ionisation laser assistée par matrice. L’analyse d’image des cartes appariées a permis d’identifier sept protéines qui étaient soit surexprimées, soit sous-exprimées : le récepteur de la protéine kinase C activée, la tubuline alpha (×2), la prostaglandine F2-alpha synthase, la protéine disulfide isomérase, une protéine du transport vésiculaire et une protéine hypothétique. L’étude souligne l’utilité de la protéomique dans l’identification des protéines pouvant être différentiellement exprimées selon le caractère sensible ou résistant des souches L. tropica.
1Department of Medical Parasitology and Mycology, School of Public Health, Tehran University of Medical Sciences, Tehran, Islamic Republic of Iran (Correspondence to H. Hajjaran:
2Protein Chemistry Unit, Biotechnology Research Centre, Pasteur Institute of Iran, Tehran, Islamic Republic of Iran.
3Department of Medical Parasitology and Mycology, School of Medicine, Tehran University of Medical Sciences, Tehran, Islamic Republic of Iran.
Received: 28/07/10; accepted: 06/10/10
EMHJ, 2012, 18 (2): 165-171
Introduction
With an annual rate of 1.5 to 2 million new cases throughout the world cutaneous leishmaniasis (CL) remains a serious public health problem in numerous countries [1,2]. From 13% to 25% of CL cases in the Islamic Republic of Iran are anthroponotic cutaneous leishmaniasis (ACL) caused by Leishmania tropica [3] and in recent years the reported number of ACL cases has increased in most parts of the country [4]. Since there is no effective vaccine against leishmaniasis, early diagnosis and appropriate treatment of patients are the best disease control measures in ACL [5]. Although the pentavalent antimonial (SbV)-based drug Glucantime® is the first choice for treatment and control of ACL, the incidence of SbV-resistant Leishmania spp. is increasing in several parts of the world [6,7], including the Islamic Republic of Iran [8]. Current data based on epidemiology and transmission studies of ACL foci of L. tropica in the north and north-east of the country have documented rates of acquired antimony drug resistance reaching 40% [3,8].
In spite of many studies, the mechanism of Leishmania antimonial resistance is still unclear [9–17]. Molecular methods, based on DNA, RNA and protein assays, have been used successfully in vivo and in vitro [9] to study resistance to anti-Leishmania drugs in different Leishmania spp [10]. Gene amplification studies have implicated various mechanisms and several genes that are over- or down-expressed. Changes in proteins profiles have been less well studied. Recently, however, comparative proteomics analysis based on 2-dimensional (2D) gel electrophoresis and mass spectrometry has been shown to be a powerful approach for determining the differences in protein patterns of subjects with leishmaniasis [17].
Our preliminary study using 2D gel electrophoresis in drug sensitive/resistant strains of L. tropica showed that some proteins were differentially expressed [18]. In the present study, we used a comparative approach with matrix-assisted laser desorption/ionization time of flight (MALDI-Tof-Tof) mass spectrometry to identify more of the proteins in clinical field isolates of promastigote forms of genetically-confirmed drug-sensitive and drug-resistant L. tropica parasites [8].
Methods
Study design
This comparative proteomics study used Sb(V)-sensitive and -resistant L. tropica strains with the aim of identifying any proteins with differential expression in drug-resistant isolates compared with drug-sensitive isolates. Three biological replicates were prepared by 3 distinct culture batches of the promastigotes in both isolates. Independent 2D electrophoresis gel samples were run and analysed using the ImageMaster software package. Differences greater than 2-fold were identified as valid protein spots by the software and these spots were excised from the gels and sent for identification by MALDI-Tof-Tof mass spectrometry.
Parasites
The 2 L. tropica strains used had been genetically confirmed previously by standard in vivo and in vitro genetic assays as either resistant (Msh-R878) or sensitive (Msh-S2) to meglumine antimoniate [8]. The isolates were recovered from Iranian patients from Mashhad city, in north-eastern Islamic Republic of Iran.
Cell culture
The sensitive and resistant L. tropica isolates were recovered from liquid nitrogen, and sub-cultured in RPMI1640 medium (Gibco/BRL) supplemented with 10% fetal bovine serum (Gibco/BRL). Cultures were incubated at 25 °C. Promastigotes at late log phase (about 400 × 106/mL) were harvested by centrifugation at 2500× g and washed 3 times with sterile phosphate-buffered saline (pH 7.2–7.4).
Sample preparation (protein extraction)
The cells were thawed and resuspended in 5 mM Tris–HCl, pH 7.8, containing 1 mM phenylmethylsulfonyl fluoride as a protease inhibitor. The samples were sonicated at 40 Hz 3 times for 10 s with 50 s intervals on ice bath. The homogenate was kept at 4 °C for 4 h. Proteins were precipitated by 20% trichloroacetic acid in acetone with 20 mM dithiothreitol (DTT) for 1 h at –20 °C. The samples were then centrifuged at 13 000× g for 15 minutes at 4 °C and the pellets were washed with cold acetone containing 20 mM DTT. Residual acetone was removed by airdrying. The pellets were resolubilized in 50 µL of sample lysis buffer containing 7M urea, 2M thiourea, 1% ampholyte pH 4–6.5, 1% ampholyte pH 5–7 and 4% CHAPS detergent. Protein concentration was determined by Bradford assay using bovine serum albumin as standards [19].
Two-dimensional gel electrophoresis
Isoelectric focusing was performed on 17 cm immobilized pH gradient (IPG) strips (BioRad) with pH range of 4–7. IPG strips were rehydrated overnight by loading the samples diluted with rehydration buffer containing 8 M urea, 4% CHAPS, 2% ampholyte, 50 mM DTT, and traces of bromophenol blue. Isoelectric focusing was carried out using the Protean IEF cell (BioRad) beginning with a linear increase from 0 to 250 V for 20 min, followed by linear increase to 10 000 V, and remaining at 10 000 V to achieve total 60 000 V/h. After focusing, IPG strips were equilibrated for 15 min. in the equilibration solution (50 mM tris–HCl pH 8.8, 6 M urea, 20% glycerol, 2% sodium dodecyl sulphate (SDS), and 0.01% bromophenol blue) containing 2% DTT, and then reacted for a further 15 min. in the equilibration solution containing 2.5% iodoacetamide. The equilibrated strips were placed on top of 10%–15% gradient SDS polyacrylamide gel electrophoresis (SDS-PAGE) slab gels and sealed with 1% agarose. The second dimension electrophoresis was performed at 16 mA/gel for 30 min. and 24 mA/gel for 5 h at 20 °C. The 2D gels were stained with mass-spectrometry-compatible silver nitrate staining [20].
Image analysis
Gel images were scanned at a resolution of 300 dpi using the BioRad GS-800 densitometer. Spot detection and matching were performed using the ImageMaster 2D Platinum software (GE Healthcare) on silver-stained analytical gels. Statistical analysis of protein variations was carried out using the Student t-test with a confidence level of 95% on relative volume of matched spots.
Mass spectrometry analysis
The protein spots were manually cut from 2D gels and destained by 50% acetonitrile/100 mM ammonium bicarbonate. Trypsin (2 mg/mL in 25 mM ammonium bicarbonate) digestion was performed at 37 °C overnight. Extracted peptides were spotted onto a 192-well MALDI-TOF target plate for the proteomics analyser (Applied Biosystems 4700), performing peptide mass fingerprinting (PMF) and subsequent tandem (MS/MS) analysis on up to 10 precursor peptides. The PMF and MS/MS results were automatically compared with the L. major genome database using the Mascot search engine (Matrix Science). Mass tolerance settings of 1.2 Da for the parent ion and 0.5 Da for fragment ions were applied. Search settings allowed 1 missed cleavage with trypsin and 2 modifications (carboxamidomethylation of cysteine and oxidation of methionine). Statistical confidence limits of 95% were applied for protein identification.
Results
The protein spots in the range of pH 4–7 were analysed and are shown in Figure 1 . There was an obvious similarity in protein spot distribution between the 2 isolates. A number of protein spots of interest in addition to the differentially expressed proteins were identified by mass spectrometry. Some of these proteins were structural proteins including alpha- and beta-tubulin, stress proteins such as heat shock protein (HSP) 70, oxidoreductase, peroxidase such as tryparedoxine peroxidase and other proteins with unknown functions such as hypothetical proteins (Table 1 ).
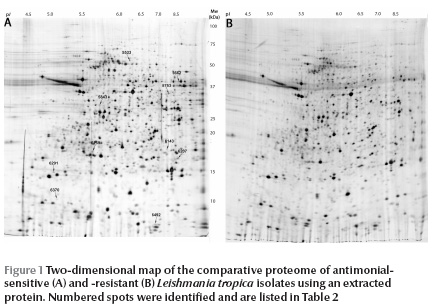
The next step of the study was to identify a number of these proteins to serve as differentials. Image analysis of the matched maps showed 7 differentially expressed proteins with significant alteration (t > 2.57) in normalized volume (Figure 2). These were 5 protein spots which were down-expressed and 2 which were over-expressed in drug-resistant isolates (Table 2 , Figure 2).
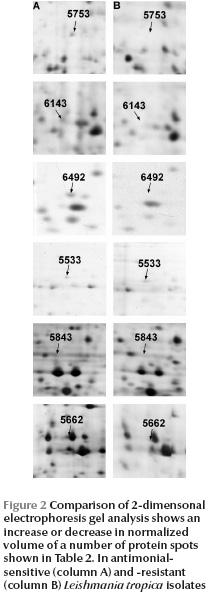
The proteins, which were highly expressed under stress conditions or drug resistance in L. tropica isolates, were identified as activated protein kinase C receptor (LACK) and a hypothetical protein. The down-expressed proteins included prostaglandin f2-alpha synthase (PGFS), protein disulfide isomerase-2 (PDI-2), alphatubulin and vesicular transport proteins (CDC48 homologue).
Discussion
The first choice chemotherapy for different leishmaniases is pentavalent antimony [10]. Recent progress in the completion of Leishmania spp. genomic sequences and access to these data permit scientists to co-study the expression of genes and proteins by powerful proteomics approach. Studying the proteome helps us to understand the differences in protein expression between Leishmania parasites reactive to and resistant to drugs [21].
The protein map of promastigote forms of the 2 strains was prepared and analysed for differences in gene expression. Each map contained about 1000 protein spots. Previous studies revealed about 2000 protein spots in L. tropica [18], 3700 in L. major [22], 2000 in L. donovani [23] and 719 in L. guyanensis [24]. The image analysis of matched maps of 2 sensitive/resistant field isolates showed 7 proteins with significant alteration in normalized expression in promastigotes. These landmark proteins were identified by mass spectrometry as LACK, alpha-tubulin, PGFS, PDI-2, vesicular transport protein and some proteins identified as hypothetical proteins.
LACK was one of the over-expressed proteins in resistant L. tropica isolates. Studies showed that production of LACK is the immune response in susceptible BALB/c mice against Leishmania and remains a good candidate for a vaccine for human leishmaniasis [25]. This protein is also required for parasite viability and represents a potential drug target for the treatment of leishmaniasis [26].
Other landmark proteins found were structural proteins including alpha-tubulin; both alpha- and gamma-tubulin are important for cell shape, growth and differentiation and for inhibition of cellular proteolysis [27]. Decreased expression levels of these proteins are probably related to reduced protection against antimonial drugs in resistant isolates.
Vesicular transport proteins identified as putative proteins were other proteins down-expressed in our resistant L. tropica strains. These proteins are located close to the flagellar pocket. Down-expression of these transport proteins decreases the transport and therefore the concentration of antimonial drugs within the cell [28].
We also report here identification of a 54 kDa protein which was shown to be PDI. Expression of PDI has been shown to result in attenuation of cell viability in response to hypoxia and to protect cells from apoptotic cell death in response to drug resistance in in vivo conditions [29]. Down-expression of this protein may be associated with parasite survival.
PGFS with 32 kDa molecular weight was another differentiation protein in field isolates of sensitive/resistant L. tropica. The PGFS protein exhibits 99.3% identity in cytosol of promastigotes in Old World Leishmania spp. in L. donovani and L. tropica [30].
A number of proteins that were over-expressed in resistant isolates were identified as hypothetical proteins, but their function remains unknown. The current sequencing data of protozoan parasites suggest that about 60% of the putative gene products are hypothetical and have no known homologues [21].
Other proteomic studies of drug resistance on promastigotes of L. donovani field Indian isolates against Sb(V) reported differential expression of at least 2 proteins. The HSP83 and a small kinetoplastid calpain-related protein were shown to be increased through modification of the programmed cell death in the resistant parasite [17]. Another study showed that in methotrexate-resistant Leishmania isolates some proteins, such as pteridine reductase (PTR1) and trypanothione reductase, were post-translationally modified and over-expressed. These proteins could allow the Leishmania parasite to quickly adjust its response to the oxidative pathway. However, the role of PTR1 in drug-resistant Leishmania spp. is not clear [31].
Conclusion
In conclusion, proteomics seem to be a potential method to differentiate protein expression in Sb(V)-resistant L. tropica strains compared with sensitive isolates. Over-expression of LACK and some hypothetical proteins as well as down-regulation of PGFS, PDI-2 and vesicular transport protein in drug-resistant strains were identified. These altered proteins seem to play a role in the drug stress response, perhaps allowing the Leishmania parasite to quickly adjust its response to the drug and delay cell death. Identification of more proteins related to the resistance mechanism requires further studies involving other technical methods.
References
- Control of the leishmaniases. Report of a WHO Expert Committee. World Health Organization Technical Report Series, 1990, 793:1–158.
- Desjeux P. Worldwide increasing risk factors for leishmaniasis. Medical Microbiology and Immunology, 2001, 190:77–79.
- Mohebali M et al. Comparison of miltefosine and meglumine antimoniate for the treatment of zoonotic cutaneous leishmaniasis (ZCL) by a randomized clinical trial in Iran. Acta Tropica, 2007, 103:33–40.
- Hajjaran H et al. Identification of Leishmania species isolated from human cutaneous Leishmaniasis, using RAPD-PCR. Iranian Journal of Public Health, 2004, 33:8–15.
- Davies CR et al. Leishmaniasis: new approaches to disease control. BMJ (Clinical Research Ed.), 2003, 326:377–382.
- Lira R et al. Evidence that the high incidence of treatment failures in Indian Kala-Azar is due to the emergence of antimony -resistant strains of Leishmania donovani. Journal of Infectious Diseases, 1999, 180:564–567.
- Rojas R et al. NG. S. Resistance to antimony and treatment failure in human Leishmania (Viannia) infection. Journal of Infectious Diseases, 2006, 15:1375–1383.
- Hadighi R et al. Unresponsiveness to Glucantime treatment in Iranian cutaneous leishmaniasis due to drug-resistant Leishmania tropica parasites. PLoS Medicine, 2006, 3:e162.
- Croft SL, Brun R. In vitro and in vivo models for the identification and evaluation of drugs active against Trypanosoma and Leishmania. In: Fairlamb AH, Ridley R, Vial H, eds. Drugs against parasitic diseases: R&D methodologies and issues. Geneva, World Health Organization, 2003:165–175.
- Alvar J, Croft S, Olliaro P. Chemotherapy in the treatment and control of leishmaniasis. Advances in Parasitology, 2006, 61:223–274.
- Berman JD, Chulay DJHD, Oster CN. Susceptibility of clinically sensitive and resistant Leishmania to pentavalent antimony in vitro. American Journal of Tropical Medicine and Hygiene, 1982, 31:459–465.
- Berman JD. Gallalee JV ABG. Sodium stibogluconate (Pentostam) inhibition of glucose catabolism via the glycolytic pathway, and fatty acid beta-oxidation in Leishmania mexicana amastigotes. Biochemical Pharmacology, 1987, 36:197–201.
- Callahan HL, Beverley SM. A member of the aldoketo reductase family confers methotrexate resistance in Leishmania. Journal of Biological Chemistry, 1992, 267:24165–24168.
- Papadopoulou B, Roy G, Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO Journal, 1992, 11:3601–3608.
- Croft SL, Sundar S, Fairlamb AH. Drug resistance in leishmaniasis. Clinical Microbiology Reviews, 2006, 19:111–126.
- Guimond C et al. Modulation of gene expression in Leishmania drug resistant mutants as determined by targeted DNA microarrays. Nucleic Acids Research, 2003, 31:5886–5896.
- Vergnes B et al. A proteomics screen implicates HSP83 and a small kinetoplastid calpain-related protein in drug resistance in Leishmania donovani clinical field isolates by modulating drug-induced programmed cell death. Molecular and Cellular Proteomics, 2007, 6:88–101.
- Hajjaran H et al. Protein profiling on meglumine antimoniate (glucantime (r)) sensitive and resistant L. tropica isolates by 2-dimentional gel electrophoresis: a preliminary study. Iranian Journal of Parasitology, 2009, 4:8–14.
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 1976, 72:248–254.
- Shevchenko A et al. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry, 1996, 68:850–858.
- Papadopoulou B, Drummelsmith JOM. Proteomics to explore pathogenesis and drug resistance mechanisms in protozoan parasites. In: Hondermarck H. ed. Biomedical and pharmaceutical applications of proteomics. New York, Kluwer Academic, 2004.
- Drummelsmith J et al. Differential protein expression analysis of Leishmania major reveals novel roles for methionine adenosyltransferase and S-adenosylmethionine in methotrexate resistance. Journal of Biological Chemistry, 2004, 279:33273–33280.
- El Fakhry Y, Ouellette M, Papadopoulou B. A proteomic approach to identify developmentally regulated proteins in Leishmania infantum. Proteomics, 2002, 2:1007–1017.
- Acestor N et al. Establishing two dimensional gels for the analysis of Leishmania proteomes. Proteomics, 2002, 2:877–879.
- Ahmed SB et al. A comparative evaluation of different DNA vaccine candidates against experimental murine leishmaniasis due to L. major. Vaccine, 2004, 22:1631–1639.
- Kelly BL, Stetson DBaL, Richard M. Leishmania major LACK antigen is required for efficient vertebrate parasitization. Journal of Experimental Medicine, 2003, 198(11):1689–1698.
- Ashutosh SS, Goyal N. Molecular mechanisms of antimony resistance in Leishmania. Journal of Medical Microbiology, 2007, 56:143–153.
- Leprohon P et al. Modulation of Leishmania ABC protein gene expression through life stages and among drug-resistant parasites. Eukaryotic Cell, 2006, 5:1713–1725.
- Zai A et al. Cell-surface protein disulfide isomerase catalyzes transnitrosation and regulates intracellular transfer of nitric oxide. Journal of Clinical Investigation, 1999, 103:393–399.
- Kabututu Z et al. Prostaglandin production from arachidonic acid and evidence for a 9,11-endoperoxide prostaglandin H2 reductase in Leishmania. International Journal for Parasitology, 2002, 32:1693–1700.
- Drummelsmith J et al. Proteome mapping of the protozoan parasite Leishmania and application to the study of drug targets and resistance mechanisms. Molecular and Cellular Proteomics, 2003, 2:146–155.


