Research article
M. Nateghpour,1 H. Abed Khojasteh,1 H. Keshavarz,1 H. Hajjaran,1 Gh. Edrissian,1 A. Rahimi 2 and N. Gobakhloo1
مقارنة بين الفحص المجهري وبين التفاعل السلسلي للبوليميراز المركَّب الجزئي التعشيش في تشخيص المتصوِّرات المنجلية والمتصوِّرات النشيطة
مهدي ناطق بور، هدى عابد خجسته، حسين كشاورز، هما حجاران، غلام حسين إدريسيان، عباس رحيمي، نفيسة قباخلو
الخلاصـة: قد قارن الباحثون في هذه الورقة بين الفحص المجهري وبين المقايسة بالتفاعل السلسلي للبوليميراز المركَّب الجزئي التعشيش، في المناطق الموبوءة من جمهورية إيران الإسلامية. وقد شملت الدراسة 68 شخصاً دماؤهم إيجابية للملاريا وهم يُبدون أعراضاً توحي بالملاريا. وفحصت أفلام كثيفة للدم بتلوين غيمزا بالمجهر الضوئي بحثاً عن طفيليات الملاريا في ساحة تكبير 100 و 200 . واستخلص الدنا من عينات الدم وطبق عليه التفاعل السلسلي للبوليميراز المركّب الجزئي التعشيش مع تضخيم متواليات جين الرنا الريبوزومي في الوحدات الفرعية الصغيرة. وأوضح الفحص المجهري % أن 48.5 % من العينات ) 33.8 % من عينات المتصورات النشيطة و 14.7 % من عينات المتصورات المنجلية( إيجابية في فحص ساحة التكبير 100 ، وأن 50 من العينات ) 35.5 % من عينات المتصورات النشيطة و 14.7 % من عينات المتصورات المنجلية( إيجابية في ساحة التكبير 200 . وأظهرَتْ نتائج التفاعل . السلسلي للبوليميراز المركّب الجزئي التعشيش، أنها مشابهة للفحص المجهري في ساحة التكبير
ABSTRACT: We compared light microscopy examination and a semi-nested multiplex PCR (SnM-PCR) assay in endemic areas of the Islamic Republic of Iran. A total of 68 individuals with malaria-positive and suspected malaria symptoms were included in the study. Giemsa-stained thick blood films were examined under a light microscope for malaria parasites in 100 and 200 fields. DNA was extracted from blood samples and SnMPCR based on the amplification of the small sub-unit ribosomal RNA (ssrRNA) gene sequences was applied. Microscopical examination showed that 48.5% (33.8% P. vivax and 14.7% P. falciparum) and 50% (35.3% P. vivax and 14.7% P. falciparum) of the samples were positive in 100 and 200 fields respectively. SnM-PCR showed the same results as the 200 field microscopy.
Comparaison de l’examen microscopique et de l’amplification génique multiplex semi-nichée pour le diagnostic de Plasmodium falciparum et P. vivax
RÉSUMÉ: Nous avons comparé l’examen microscopique optique et l’amplification génique (PCR) multiplex semi-nichée dans des zones d’endémie en République islamique d’Iran. Au total, 68 personnes dont les résultats d’analyse du paludisme étaient positifs ou qui présentaient des symptômes suspects de paludisme ont été incluses dans l’étude. Les gouttes épaisses colorées au Giemsa ont été examinés au microscope optique à la recherche de parasites du paludisme sur 100 et 200 champs microscopiques. L’ADN a été extrait des échantillons de sang et la PCR multiplex semi-nichée reposant sur l’amplification des séquences géniques d’une petite sous-unité d’ARN ribosomique a été utilisée. L’examen microscopique a révélé que 48,5 % (33,8 % pour P. vivax et 14,7 % pour P. falciparum) et 50 % (35,3 % pour P. vivax et 14,7 % pour P. falciparum) des échantillons étaient positifs sur 100 et 200 champs respectivement. La PCR multiplex semi-nichée a produit des résultats identiques à la microscopie sur 200 champs.
1Department of Medical Parasitology and Mycology;
2Department of Epidemiology and Biostatistics, School of Public Health, Tehran University of Medical Sciences, Tehran, Islamic Republic of Iran (Correspondence to H. Abed Khojasteh:
EMHJ, 2011, 17(1): 51-55
Introduction
According to the latest report released by the World Health Organization, 247 million people were affected with malaria in the world and nearly one million deaths due to the disease were recorded in 2006 [1]. Four species, Plasmodium falciparum, P. vivax, P. malariae and P. ovale are causative agents for malaria in humans. Drug selection for an effective and suitable treatment of malaria depends on precise laboratory identification of the species, and conventional light microscopy detection in Giemsa-stained thick and thin blood films is still considered the gold standard method [2]. Differentiation among species and observation of morphological changes caused by recent treatment are some advantages of the microscopy method [3]. Although this procedure is cheap and simple, it is time consuming and requires a well trained microscopist. Moreover, in cases of low parasitaemia and misdiagnosis in mixed infection, the risk of severe disease may be increased, particularly in falciparum malaria [4–6]. Therefore, in recent years many alternative approaches such as rapid diagnostic tests (RDTs) and polymerase chain reaction (PCR) methods have been developed for detection and identification of malaria parasites [7–9]. These procedures have some advantages as well as a few disadvantages [9]. PCR-based methods are not able to evaluate parasite morphology through the developmental stages [10].
This study was conducted to compare semi-nested multiplex (SnM)-PCR and the microscopical method in 100 and 200 fields of thick blood films for diagnosis of malaria parasites. The examination of 100 oil immersion fields is usually a standard criterion for routine purposes of malaria parasite identification [11]. Examination of 200 oil immersion fields is recommended in areas with relatively low degrees of parasitaemia [12].
Methods
Study area
This study was undertaken from November 2006 to July 2007 in the Minab and Bandar-Abbas districts in Hormozgan province and Iranshahr district in Sistan and Baluchistan province, both malarious provinces, located in the south and south-eastern part of the Islamic Republic of Iran respectively.
The weather in these provinces is usually warm and humid. Nearly 80% of the detected cases of vivax and falciparum malaria come from these parts of the country (Annual report of Centre for Disease Management in Iran ). P. vivax is the predominant species and P. falciparum the second commonest in these areas.
Sample collection
We selected 3 health centres as passive sentinel sites for sampling in each district. A total of 68 individuals (56 from Hormozgan, 12 from Sistan and Baluchistan provinces) aged 2–60 years, comprising 18 females (26.5%) and 50 males (73.5%), were enrolled in the study: 34 of the considered cases were microscopical malaria positive and the other 34 cases were selected with suspected symptoms of malaria to optimize the SnM-PCR method. At the same time, before treatment, 2 Giemsa-stained thick films were made from each patient and venepuncture blood was collected in tubes containing EDTA for microscopical examination and SnM-PCR assay (1 mL for each). The venepuncture blood samples were stored at –20 °C until DNA extraction could be carried out.
The number of samples needed was calculated based on 11% disagreement between PCR and microscopical examination for diagnosis of P. falciparum and P. vivax [13] with 95% confidence level and 80% power [14].
Microscopy
Thick blood films were air-dried and stained for 30 min in 3% diluted Giemsa stain (pH 7.2). The slides were then rinsed with tap water and air-dried.
Thick films were diagnosed by skilled local microscopists at the place of sampling. All the slides were examined under × 1000 magnification for 100 and 200 oil immersion fields. In line with the aim of this study, a plus system of counting was used to determine parasitaemia: a code of 1–4 plus signs (+) was used [15] as follows:
+ = 1–10 parasites per 100 thick film fields,
++ = 11–100 parasites per 100 thick film fields,
+++ = 1–10 parasites per single thick film field,
++++ = > 10 parasites per single thick film field.
Thick blood films and blood samples were sent to the Department of Medical Parasitology in Tehran University of Medical Sciences to confirm the parasite species by PCR analysis and double-checking of microscopical examination.
DNA extraction
DNA was extracted from 200 μL EDTA whole blood with the FlexiGene DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. DNA samples were stored at –20 °C until PCR could be carried out.
SnM-PCR was performed with primers described previously [13], with some modifications. The primers that were used, (DNA-Technology, Aarhus, Denmark) comprised a reverse primer (UNR) (5′-GACGGTATCTGATCGTC TTC-3′) that would hybridize to all Plasmodium spp. and a wide variety of vertebrates, 2 forward primers that hybridize to mammals (HUF) (5′-GAGCCGCCTGG ATACCGC-3′) or to all Plasmodium spp. (PLF) (5AGTGTGTATCAATCGAGTTTC-3′) and 2 other reverse primers (FAR) (5′-AGTTCCC CTAGAATAGTTACA-3′) hybridize only to P. falciparum and (VIR) (5′-AGG ACTTCCAAGCCGAAGC-3′) which hybridizes only to P. vivax.
PCR amplification
This method has 2 main steps: a first amplification with specific Plasmodium primers for ssrDNA and a second amplification having 2 separate amplifications with independent pairs of primers for P. vivax and P. falciparum [13]. In the first reaction, using UNR, HUF, and PLF primers, we expected to have 2 products: a 231 bp band from UNR-HUF (the positive control for all samples), and a band of 787 bp resulting from UNR-PLF which shows the presence of any Plasmodium spp.; this band detected only parasitaemia of more than 3% (Figure 1).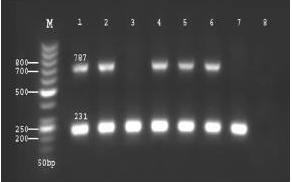 3% parasitaemia; lane 3: Plasmodium-infected sample with
3% parasitaemia; lane 3: Plasmodium-infected sample with
Figure 1 Agarose gel electrophoresis of amplification products of first reaction of SnM-PCR [lane M: 50 bp marker DNA ladder; lanes 1, 2, 4, 5, 6: Plasmodium-infected samples with > 3% parasitaemia; lane 3: Plasmodium-infected sample with
In the second reaction, which identifies Plasmodium spp., the PLF, FAR and VIR primers were used. A band of 395 bp with PLF-FAR indicates P. falciparum infection and a band of 499 bp with PLF-VIR indicates P. vivax infection (Figure 2).![Figure 2 Agarose gel electrophoresis of amplification products of second reaction of SnM-PCR [lane M: 50 bp marker DNA ladder; lanes 1–8: Plasmodium falciparum (395bp); lanes 9–12: Plasmodium vivax (499 bp); lane 13: negative control]](/images/stories/emhj/vol17/01/articl33.jpg)
Figure 2 Agarose gel electrophoresis of amplification products of second reaction of SnM-PCR [lane M: 50 bp marker DNA ladder; lanes 1–8: Plasmodium falciparum (395bp); lanes 9–12: Plasmodium vivax (499 bp); lane 13: negative control]
The first amplification reaction was performed in volumes of 50 µL and 2 µL of isolated DNA that were added to a PCR Master Mix, containing 1.75 mM MgCl2, 200 µM dNTPs, 25, 25 and 10 pmol of UNR, PLF and HUF primers respectively, and 1.0 U of Taq polymerase (Roche Biotech) in the PCR buffer. The second PCR reaction was performed in 25 µL and 2 µL of 1/500 diluted PCR product from the first reaction, which were added to a PCR Master Mix containing 1.75 mM MgCl2, 200 µM dNTPs, 25, 10, 10 pmol of PLF, FAR and VIR primers respectively and 0.5 U of Taq polymerase (Roche Biotech) in the PCR buffer. Reactions were overlaid with 25 µL mineral oil. A thermal cycler (Techne, United States of America) was used for both reactions with some modifications in the cycling programmes. Reaction conditions (first reaction) were denaturation at 94 °C for 4 min. followed by 35 cycles of 94 °C for 45 s, 48 °C for 1.5 min and 72 °C for 2 min. plus an extension at 72 °C for 8 min. The second PCR condition followed with initial denaturation at 94 °C for 3 min. and 35 cycles of 94 °C for 30 s, 48 °C for 30 s and 72 °C for 1 min plus an extension at 72 °C for 5 min. PCR products were stained with ethidium bromide for visual detection by ultraviolet transillumination, and resolved by 1% agarose gel electrophoresis for both reactions.
Genomic DNA from healthy individuals, with no history of malaria, living in non-endemic areas of the country were prepared and used as negative controls in all PCR assays. A P. berghei DNA sample was also used as the positive mammalian control, which gave a UNR–HUF band on the first amplification but no band on the second one.
Results
The comparative diagnosis results of P. vivax and P. falciparum between SnM-PCR procedure and microscopic method are summarized in Tables 1 and 2. Conventional microscopy on 100 fields of thick blood smears identified 23 (33.8%) of the cases as P. vivax and 10 (14.7%) as P. falciparum (Table 1). In contrast 24 (35.3%) were identified as P. vivax and 10 (14.7%) as P. falciparum on 200 fields of thick blood smears (Table 2). Using SnM-PCR the same results were obtained as for 200 fields of microscopical examination. To demonstrate the parasitaemia situation, the results for counting parasites in 100 thick smear fields are tabulated in Table 3. Meanwhile, 1 thick smear of P. vivax which was negative in 100 fields became 1+ when counting was carried out on up to 200 fields.
Table1 Comparative identification study between SnM-PCR and microscopical method (100 fields of thick blood smear) for diagnosis of Plasmodium falciparum (Pf) and P. vivax (Pv)
Table 2 Comparative identification study between SnM-PCR and microscopical method (200 fields of thick blood smear) for diagnosis of Plasmodium falciparum (Pf) and P. vivax (Pv)
Table 3 Number of parasites in 100 thick smear fields of Plasmodium vivax and P. Falciparum
McNemar analysis showed significant agreement between the results for the SnM-PCR method and 100 fields (kappa = 0.97, P = 1.00) and also 200 fields (kappa = 1.00, P = 1.00) microscopical examination.
In order to confirm the SnM-PCR results and the specificity assurance of this method, 3 isolates were randomly sequenced by MWG Biotech (Ebersberg, Germany) and submitted in the National Center for Biotechnology Information (NCBI) gene bank databases as: IRN-BV1 (accession number: EU551666, P. vivax from Bandar-Abbas district, Hormozgan province), IRN-SF1 (accession number: EU551667, P. falciparum from Sistan and Baluchistan province), IRN–BF1 (accession number: EU551668 P. falciparum from Bandar-Abbas district, Hormozgan province)
Discussion
Since the latter part of the 19th century the microscope has been used as an useful tool for the detection of P. falciparum [16] and it is still considered the best tool for the diagnosis of malaria parasites in most malarious areas. The emergence of novel and sensitive techniques based on molecular studies such as PCR means the accuracy of light microscopic diagnosis of malaria parasites has become more or less negotiable by some researchers.
Results from a study in a malaria-endemic but unstable transmission district showed that 158, 89, 8 and 25 out of 280 Giemsa-stained slides examined with light microscope were P. vivax, P. falciparum, mixed (P. falciparum and P. vivax) and negative cases respectively [17]. Using nested PCR assay for the same blood samples resulted in 145, 79, 41 and 15 P. vivax, P. falciparum, mixed and negative respectively The same pattern of results was found in another Iranian study [18], i.e. PCR lowered the detection of P. falciparum and P. vivax positive cases compared with microscopy. Another comparative study showed a pattern of results different from these 2 for P. vivax and P. falciparum, i.e. PCR increased the detection of positive cases: out of 174 Giemsa-stained slides examined with the light microscope 40 were P. falciparum, and 26 were P. vivax; using nested PCR assay on the same samples, 50 were P. falciparum and 31 P. vivax [2].
Tham et al. showed that out of 52 samples 16, 34 and 0 were diagnosed as P. falciparum, P. vivax and mixed respectively by light microscopy and 16, 34, 2 were diagnosed as P. falciparum, P. vivax and mixed respectively by PCR [7]. PCR assay was considered equivalent to light microscopy method in the diagnosis of P. falciparum and P. vivax.
In the present study we compared PCR technique with conventional microscopy. Our findings showed that there were no significant differences between PCR assay and microscopical examination for malaria diagnosis if the thick blood smears were examined precisely in at least 100 fields by a trained microscopist. The results of microscopical examination of 200 fields were closer to the results of PCR than 100 fields of thick blood films. In fact, microscopical diagnosis of malaria parasites depends primarily on the skill of the personnel and the quality of the materials and equipment.
In conclusion, although PCR is a valuable method for detecting malaria parasites, particularly in low-level parasitaemia, microscopical diagnosis still retains its unique usefulness for detection of malaria parasites, particularly in rural areas where the PCR technique is not available. Moreover, microscopy in combination with PCR can generate a more reliable approach to identifying malaria parasites when there is a need for more specific results in a particular situation.
Acknowledgements
The authors wish to thank S. Rezayee, Gh. Mohseni, E. Torabi, M. Yareian, A. Amiri, M.T. Satvat and S. Charehdar for their cooperation in this study.
This investigation received technical and financial support from Tehran University of Medical Sciences.
References
- World Malaria Report, 2008. Geneva, World Health Organization, 2008 (http://malaria.who.int/wmr2008/malaria2008.pdf, accessed 22 September 2010)
- Johnston SP et al. PCR as a confirmatory technique for laboratory diagnosis of malaria. Journal of Clininical Microbiology, 2006, 44:1087–1089.
- Di santi SM et al. PCR-Based diagnosis to evaluate the performance of malaria reference centers. Revista do Instituto de Medicina Tropical de São Paulo, 2004, 46:183–187.
- Perandin F et al. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. Journal of Clininical Microbiology, 2004, 42:1214–1219.
- Speer DJ et al. Diagnosis of malaria aided by polymerase chain reaction in two cases with low-level parasitaemia. Internal Medicine, 2003, 3:613–515.
- Hänscheid T. Diagnosis of malaria: a review of alternatives to conventional microscopy. Clinical & Laboratory Haematology, 1999, 21:235–245.
- Tham JM et al. Detection and species determination of malaria parasites by PCR: comparison with microscopy and with para sight – F and ICT malaria pf tests in a clinical environment. Journal of Clinical Microbiology, 1999, 37:1269–1273.
- Singh B et al. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. American Journal of Tropical Medicine and Hygiene, 1999, 60:687–692.
- Malaria diagnosis, new perspectives. Report of a joint WHO/USAID informal consultation. Geneva, World Health Organization, 2000.
- Karl S et al. Enhanced detection of gametocytes by magnetic deposition microscopy predicts higher potential for Plasmodium falciparum transmission. Malaria Journal, 2008, 7:66 (http://www.malariajournal.com/content/7/1/66, accessed 28 September 2010).
- Payne D.Use and limitations of light microscopy for diagnosing malaria at the primary health care level. Geneva, World Health Organization, 1988.
- Trape JF. Rapid evaluation of malaria parasite density and standardization of thick smear examination for epidemiological investigations. Transactions of the Royal Society of Tropical Medicine and Hygiene, 1985, 79:181–184.
- Rubio JM et al. Alternative polymerase chain reaction method to identify plasmodium species in human blood samples: the Semi-Nested Multiplex malaria PCR (SNM-PCR). Transactions of the Royal Society of Tropical Medicine and Hygiene, 2002, 96:199–204.
- Altman, G. Practical statistics for medical research. London, Chapman and Hall, 1991.
- Basic malaria microscopy. Geneva, World Health Organization, 1991.
- Wernsdorfer WH, McGregor I. Malaria, principles and practice of malarialogy. Harlow, Essex, Longman Group UK Limited, 1988.
- Zakeri ST et al. Detection of malaria parasites by nested PCR in south-eastern, Iran: evidence of highly mixed infections in Chahbahar district. Malaria Journal, 2002, 1(2) (http://www.malariajournal.com/content/1/1/2, accessed 28 September 2010).
- Ebrahimzadeh A, Fouladi B, Fazaeli A. High rate of detection of mixed infection of Plasmodium vivax and Plasmodium falciparum in south-east of Iran, using nested PCR. Parasitology International, 2007, 56:61–64.


