M. Abbassi,1 M. Rahbar,1 S. Hekmat Yazdi,1 F. Rashed Marandi,1 R. Sabourian1 and M. Saremi1
ABSTRACT We evaluated the performance of microbiology laboratories in the 10th run of the external quality assessment scheme (EQAS) in Tehran and districts. Each laboratory was sent 2 species of bacteria for identification. Of the 487 laboratories that participated, 437 returned their findings. While 77.0% and 69.9% correctly identified Staphylococcus saprophyticus and Citrobacter freundii respectively, only 29.8% correctly identified Acinetobacter baumanii, 25.3% identified Enterococcus faecalis and 35.6% identified Enterobacter agglomerans. However 78.7% and 79.5% of the laboratories reported correct results for susceptibility testing for S. saprophyticus and C. freundii respectively.
Évaluation des résultats du 10e programme d’évaluation externe de la qualité dans les laboratoires de microbiologie clinique à Téhéran et dans les districts
RÉSUMÉ Nous avons évalué la performance des laboratoires de microbiologie lors du 10e cycle du programme d’évaluation externe de la qualité (EQAS) à Téhéran et dans les districts. Chaque laboratoire a reçu deux espèces de bactéries pour identification. Sur les 487 laboratoires participants, 437 ont envoyé leurs résultats. Alors que 77,0 % et 69,9 % des laboratoires ont identifié correctement Staphylococcus saprophyticus et Citrobacter freundii respectivement, seuls 29,8 % ont identifié correctement Acinetobacter baumanii, 25,3 % ont identifié Enterococcus faecalis et 35,6 % ont identifié Enterobacter agglomerans. Toutefois, 78,7 % et 79,5 % des laboratoires ont fourni des résultats corrects pour les tests de sensibilité sur S. saprophyticus et C. freundii respectivement.
1Bo-Ali Hospital, Reference Laboratory of Iran, Tehran, Islamic Republic of Iran (Correspondence to M. Rahbar:
Received: 09/02/04; accepted: 25/07/04
EMHJ, 2006, 12(3-4): 300-309
Introduction
An external quality assessment scheme (EQAS) is a component of quality assurance in which laboratories participating in the programme receive EQAS specimens at regular intervals, evaluate them by routine methods and report the results to the organizing centres. In addition, EQAS is an educational tool that improves national and regional standards [1–5].
The reference laboratory of the Islamic Republic of Iran is part of the government sector. Since 1994 they have introduced EQAS in many public and private medical laboratories. In microbiology laboratories various steps have been taken to upgrade the EQAS programme. These steps have included running national training courses for trainers, sending unknown specimens at regular intervals to the laboratories of the national network and sending standardized methods to control and check the culture media and antibiotic discs.
At present, 2000 microbiology laboratories throughout the country participate in EQAS organized by the reference laboratory’s microbiology laboratory. In each trial, 2 different bacterial species are sent to the laboratories for identification (genus, species and subtype); sensitivity testing is also required for 1 of the species. The bacteria are in a pure form taken from samples sent by the World Health Organization (WHO) as part of the WHO external quality assessment scheme or bought from Difco, an American Type Culture Collection (ATCC) (part of BD Diagnostics, New Jersy, USA). Our goal is to use EQAS as an accreditation system for laboratories in the near future. In the present study we evaluated the 10th run of EQAS performance of the microbiology laboratories in Tehran and districts.
Methods
The 10th run of EQAS was carried out in July 2002 among 487 intermediate and peripheral laboratories (public and private) located in Tehran and surrounding districts. For this purpose, 5 different bacterial species (ATCC) or strains confirmed by WHO were selected and each laboratory randomly received 2 unknown samples for identification. The species were:
- Staphylococcus saprophyticus (ATCC: 15305)
- Citrobacter freundii (confirmed by WHO)
- Acinetobacter baumanii (ATCC: 19606)
- Enterococcus faecalis (ATCC: 29212)
- Enterobacter agglomerans (confirmed by WHO)
Second confirmation and other necessary biochemical tests were performed in the reference laboratory before distributing the species [2]. In addition, susceptibility testing was performed for S. saprophyticus and C. freundii using standard methods suggested by the National Committee for Clinical Laboratories Standards (NCCLS) [6]. The species then were cultured on trypticase soy agar (TSA) in screw-capped tubes. They were incubated at 35 ± 2 °C for 24 hours and then sealed. The TSA tubes, together with guidelines and necessary forms, were sent to each participating laboratory. The laboratories were asked to test one species (the S. saprophyticus) against ampicillin, tetracycline, nalidixic acid, ciprofloxacine and co-trimoxazole and the C. freundii against ampicillin, gentamicin, cephalexin, chloramphenicol and cefotaxime. They were also asked to measure the zone of inhibition using standard methods and to categorize them as susceptible (S), intermediate susceptibility (I) and resistant (R).
After identifying the species, the laboratories returned the completed forms within 1 month. The results were scored according to WHO criteria. The maximum points for complete identification of each unknown species was 3 and 5 for susceptibility testing. Each incorrect answer scored zero and a partially correct answer (doing some tests and identifying the genus) scored 0.5–2.5. If susceptibility for each antibiotic differed from the true result by one degree, 0.5 score was given [7].
Results
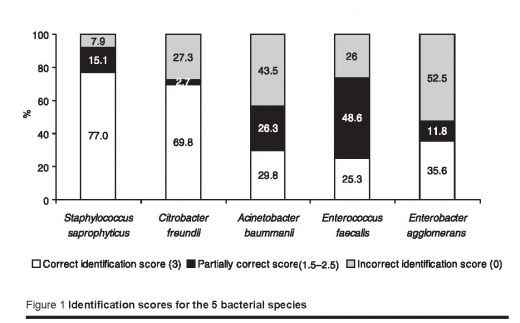
A total of 487 laboratories participated in the 10th EQAS in Tehran and districts and were sent the species for identification. Of these, 437 laboratories returned their results (89.7%). Of the 291 laboratories that were sent S. saprophyticus, 224 (77.0%) identified the organism correctly, and 102 of 146 (69.9%) laboratories correctly identified C. freundii. Of the other species, 34 of 114 (29.8%) laboratories correctly identified A. baumanii, 37 of 146 (25.3%) correctly identified Enterococcus faecalis, and 63 of 177 (35.6%) laboratories correctly identified Enterobacter agglomerans. The incorrect identification of these bacteria were 7.9%, 27.3%, 43.8%, 26.0% and 52.5% respectively (Figure 1). The mean scores and standard deviations (SD) for each bacteria were as follows:
- S. saprophyticus 2.5 (SD 0.96)
- Enterococcus faecalis 1.8 (SD 1.21)
- Enterobacter agglomerans 1.2 (SD 1.32)
- C. freundii 2.1 (SD 1.33)
- A. baumanii 1.4 (SD 1.21)
Most of the laboratories were able to perform the susceptibility testing correctly. The results were as follows: 78.7% of laboratories reported correct susceptibility testing for S. saprophyticus and 79.5% for C. freundii (Tables 1 and 2). The mean scores for susceptibility testing were 3.64 (SD 0.65) for S. saprophyticus and 4.35 (SD 1.32) for C. freundii.
Ampicillin resistance of S. sapropyticus was not reported correctly in 70% of the cases. Because of this observation, the ampicillin disks, which are produced locally, were checked and we found that the failure in identification was related to a problem with the disks themselves. Therefore, this score was omitted from the total scoring. At the end of the evaluation process, expected correct answers were sent to each laboratory. Correct identification guidelines and necessary references accompanied all the results as well.
Discussion
The main goal of EQAS is to improve the quality and strengthen the capabilities of laboratories. In evaluating the microbiology laboratories in Tehran and surrounding districts, it was presumed beforehand that the laboratories were functioning within an acceptable range. Unfortunately, our results did not confirm this assumption, and there was a wide range of capabilities of the laboratories to identify different species. This failure could have been a result of defective reagents and differential culture media, inappropriate internal quality control programmes and inadequate numbers of qualified technicians. The first external quality assessment of clinical microbiology laboratories in Norway in 1982 included 15 country and regional laboratories [8]. The mean number of incorrect identifications was 2.7 (11.3%). Eleven (11) strains were correctly identified by all laboratories, whereas 4 strains were misidentified by 4 to 7 laboratories, accounting for approximately 50% of all misidentifications [8]. According to Richardson and his associates in Canada, the number of participating microbiology laboratories in EQAS declined from 335 laboratories in 1974 to 190 laboratories in 1994 [9]. In the initial evaluation, 21% of laboratories did not have the expected capabilities. In 1989, 50% of laboratories achieved high points (above 80%) for isolating and identifying the microorganisms. However, 25% of laboratories scored less than 50% for bacterial sensitivity testing and only 10% of them had high scores (above 80%). This lack of effectiveness was related to inappropriate selection of chemicals [9].
In another study to evaluate bacterial resistance, the Centers for Disease Control and Prevention (CDC) and WHO distributed 6 different strains of bacteria among 130 laboratories in the United States and other countries. Most of the laboratories were able to identify S. aureus, Enterobacter faecalis and Klebsiella pneumoniae against methicillin, vancomycin and cephalosporin respectively [10]. However, the rest, especially those that used the disk diffusion method for evaluating the sensitivity of S. pneumoniae against penicillin, had problems. In addition, the majority of laboratories had problems evaluating reduced sensitivity of S. epidermidis to vancomycin [10]. Another study showed only 3 of 23 reference laboratories were able to identify correctly 6 lyophilized Corynebacterium diphtheriae strains and to detect the C. diphtheriae toxigenicity [11]. A study by Kumasaka in Tokyo revealed that poor performance in the EQAS survey was closely related to poor laboratory management, the type of training, experience of the medical technicians, and the supervisory ability of the consultant physicians in independent laboratories [12]. In a study in the United Kingdom, Pitt and Sands concluded that the physiological concepts of job satisfaction and climate are factors that might affect external and internal quality control [13].
We are planning to establish a proper policy for manufacturers (or importers) to produce the necessary and important media and reagents. In addition, adding special postgraduate training courses and distribution of scientific guidelines will be helpful. With these new policies, we hope in future to upgrade the capabilities of the microbio-logy laboratories in Tehran and districts.
References
- Isenberg HDE, ed. Essential procedures for clinical microbiology, 1st ed. Washington DC, American Society for Microbiology, 1998:734–5.
- Mahon C, Manuselis G. Textbook of diagnostic microbiology, 2nd ed. Philadelphia, WB Saunders, 2000:106–28.
- Koneman WEW et al. Color atlas and textbook of diagnostic microbiology, 5th ed. Philadelphia, Lippincott, 1977:113–20.
- Schal KP. The German external quality assessment scheme for bacteriology. Medical microbiology letters, 1994, 3:247–58.
- Vandepitte J et al. Basic laboratory procedures in clinical bacteriology. Geneva, World Health Organization, 1991.
- Performance standards for antimicrobial susceptibility testing. Twelfth information supplement. Wayne, Pennsylvania, NCCLS, 2002 (NCCLS document M100–12).
- Vandepotte J. External quality control in microbiology. Leven, Belgium, WHO collaborating center for external quality assessment in clinical microbiology, 1998.
- Lassen J, Sandven P. External quality assessment for clinical microbiological laboratories in Norway 1982. Evaluation of the identification of 24 bacterial strains. NIPH annals, 1983, 6:23–35.
- Richardson H et al. Quality improvement of diagnostic microbiology through a peer-group proficiency assessment program. A 20-year experience in Ontario. The Microbiology Committee. Archives of pathology & laboratory medicine, 1996, 120(5):445–55.
- Tenover FC et al. Ability of laboratories to detect emerging resistance: Proficiency testing and quality control results from the World Health Organization external assurance system for antimicrobial susceptibility testing. Journal of clinical microbiology, 2001, 39:241–50.
- Engler KH, Kozlov RS, Copping SJ. International external quality assessment schemes for the laboratory diagnosis of diphteria. Journal of medical microbiology, 2000, 50:1006–12.
- Kumasaka K. External quality assessment for clinical microbiology and good laboratory management. Rinsho byori, 1998, 46:124–31 [In Japanese].
- Pitt SJ, Sands RL. Effect of staff attitudes on quality in clinical microbiology services. British journal of biomedical science, 2002, 59:69–75.


