L. Metwally,1 N. Gomaa 1 and R. Hassan 2
الكشف المباشر للعنقوديات الذهبية المقاومة للميثيسيلين من خلال التضخيم المتساوي الحرارة بواسطة العروة، واختبارات انتشار قرص السيفوكسيتين المباشر
لبنى متولي، ناهد جمعة، رانيا حسن
الخلاصة: قد قامت الباحثات بتقييم فائدة طريقتين للكشف المباشر للعقديات الذهبية المقاومة للميثيسيلين المستمدة من زجاجة زرع الدم الإيجابية العلامة؛ والطريقتان هما المقايسة بالتضخيم المتساوي الحرارة بواسطة العروة، واختبار الانتشار المباشر لقرص وغرام. وأجرت الباحثات بموازاة ذلك اختبارات للكشف المعياري للمكروبات مع الاستجابة للأوكسيسيلين باستخدام التفاعل السلسلي للبوليمراز على مادة MecA. ومن بين 60 زجاجة زرع دم إيجابي للجراثيم المكوَّرة الإيجابية الغرام، تبيَّن أن هناك 100 % استجابة ونوعية لكشف التضخيم المتساوي الحرارة بواسطة العروة (عبر كشف جينات FemA و MecA). وعند إجراء الاختبار على المكورات العنقودية السلبية لإنزيم التخثير (كوأغيولاز) وجدت الباحثات أن حساسية كشف المقاومة للميثيسيلين بلغت 91.7 % بينما بلغت النوعية 100 % كما أن استخدام الانتشار المباشر لقرص السيفوكسيتين مع المقايسة المباشرة في الأنبوب لإنزيم التخثير لم يكشف إلا 80.6 % من الجراثيم العنقودية المقاومة للميثيسيلين، والمستجيبة له. وهكذا اتضح أن التضخيم المتساوي الحرارة بواسطة العروة يوفر دقة تشخيصية أعلى مما يوَّفره الانتشار المباشر لأقراص السيفوكسيتين، وأنه أكثر مردوداً ولا يتطلب إمدادات أو كواشف إضافية.
ABSTRACT We evaluated the utility of 2 methods for detection of methicillin-resistant Staphylococcus aureus (MRSA) directly from signal-positive blood culture bottles: loop-mediated isothermal amplification (LAMP) assay, and direct cefoxitin disk diffusion (DCDD) test using a 30 μg cefoxitin disk. In parallel, standard microbiological identification and oxacillin susceptibility testing with MecA PCR was performed. Of 60 blood cultures positive for Gram-positive cocci in clusters, LAMP (via detection of the FemA and MecA genes) showed 100% sensitivity and specificity for identification of MRSA/MSSA. When coagulase-negative staphylococci were tested, sensitivity for detection of methicillin resistance was 91.7% and specificity was 100%. DCDD along with direct tube coagulase assay detected only 80.6% of MRSA/MSSA. LAMP showed higher diagnostic accuracy although DCDD was more cost-effective and did not require additional reagents or supplies.
Détection de Staphylococcus aureus résistant à la méthicilline directement par amplification isotherme induite par boucle et par tests de diffusion sur disque à la céfoxitine directs
RÉSUMÉ Nous avons évalué l'utilité de deux méthodes de détection de Staphylococcus aureus résistant à la méthicilline directement à partir des flacons d'hémoculture donnant des signaux positifs à l'aide de l'amplification isotherme induite par boucle ainsi que de tests de diffusion sur disque de 30 μg de céfoxitine directs. En parallèle, une identification microbiologique normalisée et un test de sensibilité à l'oxacilline par PCR visant l'amplification du gène MecA ont été réalisés. Sur 60 hémocultures positives pour les cocci à Gram positif en grappes, l'amplification isotherme induite par boucle (au moyen du dépistage des gènes FemA et MecA) a montré une sensibilité et une spécificité de 100 % pour l'identification de Staphylococcus aureus résistant et sensible à la méthicilline. Lorsque les staphylocoques à coagulase négative ont été analysés, la sensibilité pour la détection de la résistance à la méthicilline était de 91,7 % et la spécificité de 100 %. Les tests de diffusion sur disque de céfoxitine directs ainsi que le dosage direct de la coagulase à partir des flacons ont détecté seulement 80,6 % des Staphylococcus aureus résistants/sensibles à la méthicilline. L'amplification isotherme induite par boucle a montré une exactitude diagnostique supérieure, même si les tests de diffusion sur disque à la céfoxitine directs étaient d'un meilleur rapport coût-efficacité et n'exigeaient ni réactifs ni fournitures supplémentaires.
1Department of Microbiology; 2Department of Clinical Pathology, Faculty of Medicine, Suez Canal University, Ismailia, Egypt (Correspondence to L. Metwally:
Received: 11/03/13; accepted: 10/10/13
EMHJ, 2014, 20(4): 273-279
Introduction
Staphylococcus aureus bacteraemia is associated with high mortality rates and prolonged hospital stays [1–3], and therefore prompt detection and differentiation of S. aureus from coagulase-negative staphylococci (CoNS) and methicillin-susceptible strains has significant prognostic, therapeutic and economic value. Fast turnaround times for positive results, defined as definitive identification of methicillin-resistant S. aureus (MRSA), are crucial so that appropriate antimicrobial agents can be chosen promptly [4]. Conventional methods for the identification of S. aureus in positive blood cultures normally require at least 24 hours, during which time patients receive empirical treatment, while an additional 24 hours is necessary to complete the antibiotic susceptibility testing [4].
Although a number of rapid and accurate tests have been developed for detection of MRSA directly from blood culture bottles flagged positive [5–10], these tests are much more costly than conventional methods, and those using molecular testing with real-time polymerase chain reaction (PCR) often require molecular expertise and equipment that may not be readily available in laboratories in developing countries. A rapid and low-cost diagnostic assay for MRSA from blood culture bottles is particularly needed in hospitals in Egypt, where wide dissemination of MRSA has been reported by previous studies [11,12]. For instance, in a study by Said et al. in 3 large university hospitals, 64% of S. aureus isolates causing bloodstream infections were MRSA [13]. The authors attributed these findings to a number of factors, including empirical treatment without laboratory confirmation of susceptibility patterns and incomplete treatment courses with antibiotics.
Loop-mediated isothermal amplification (LAMP) is a strand displacement amplification technique [14] which utilizes a set of 4 to 6 specially designed oligonucleotide primers and a specific DNA polymerase (Bst). Via the process of strand displacement amplification, a dumbbell DNA structure is produced which serves as a template for cycle amplification. The lack of a need for a thermocycler, the speed of the reaction and visual read-out make LAMP a promising platform for the development of a simple and sensitive near-patient tool for the molecular detection of MRSA in resource-limited settings [15].
In the light of the pressing need for hospitals in Egypt to implement sound infection prevention and control programmes to reduce the transmission of MRSA, this study aimed to test 2 approaches for rapid detection of MRSA directly from blood culture bottles: the LAMP method targeting the S. aureus-specific FemA and MecA genes responsible for methicillin resistance; and the direct cefoxitin disk diffusion (DCDD) test along with direct tube coagulase. These tests were compared with conventional identification assays to evaluate the usefulness of these rapid tests and to assess the appropriateness of incorporating either of these protocols into the algorithm of testing blood cultures with Gram-positive cocci in clusters (GPCC) in a peripheral laboratory.
Methods
Clinical specimens
BacT/Alert blood culture bottles (bioMérieux) were routinely inoculated with 5–10 mL blood from adult patients. Bottles were inserted into BacT/Alert 3D 60 instruments (bioMérieux) and incubated at 37 °C. When a positive signal was indicated, the culture fluid was retrieved and Gram stained.
A total of 60 consecutive blood culture bottles (1 per patient) that were positive for GPCC were collected from patients with suspected bacteraemia in Suez Canal University Hospital, Ismailia, Egypt, from June to December 2012. In addition, 15 specimens from blood culture bottles that were negative for bacterial and fungal pathogens after 5 days incubation were included as negative control samples.
Approval to perform the study was obtained from the ethics committee in the Faculty of Medicine, Suez Canal University, Egypt and the management board of the hospital.
Laboratory methods
If GPCC were observed in samples, a 1 mL aliquot was used directly for the LAMP assay and DCDD test. In parallel, standard microbiological identification and antibiotic susceptibility testing were performed.
Standard microbiological identification and antibiotic susceptibility testing
For this purpose, blood culture medium was subcultured onto 5% sheep blood agar and chocolate agar (Oxoid) and incubated at 35 °C aerobically in a 5% CO2 for 24 to 48 h. Bacterial isolates were identified primarily by colony morphology, Gram stain, catalase and tube coagulase tests. Definite identification was performed using the API Staph-Ident system (bioMérieux).
In vitro antimicrobial susceptibility testing to identify oxacillin resistance in strains was accomplished using 30 μg cefoxitin (Oxoid) disk by a disk diffusion method in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines using Muller-Hinton agar (Oxoid) and McFarland 0.5 standard [16].
Detection of the MecA gene was performed by conventional PCR methods using previously described primers [17]. Positive blood culture bottles were reincubated at 37 °C until the results of direct tests were available. S. aureus (ATCC 25923) and MRSA (ATCC 33592) were used as controls.
Direct tube coagulase and direct cefoxitin disk diffusion test
The tube coagulase test was performed directly from the blood culture broth; a positive result indicated by formation of a clot was evaluated after incubation of 5 drops of culture broth and 0.5 mL of rabbit plasma at 35 °C for 4 h [18]. The test results were reported the same day that the blood culture flagged positive. Specimens that were negative by direct-tube coagulase were subjected to slide and/or tube coagulase testing from isolated colonies carried out the next morning along with interpretation of the DCDD test.
The DCDD test was performed directly from blood culture bottles according to the method described previously [19], except for the use of standardized Muller-Hinton agar in place of tryptic-soy-blood agar plate. In brief, a sterile swab, soaked with an aliquot of blood culture fluid, was streaked on a 100 mm Muller-Hinton agar plate (Oxoid) and afterwards a cefoxitin disk (30 μg; Oxoid) was placed and the plates were incubated for 24 h at 37 °C. The zone of inhibition around the cefoxitin disk was measured and interpreted as susceptible, resistant or indeterminate compared with standardized cefoxitin disk diffusion testing using the CLSI breakpoints (for S. aureus: susceptible ≥ 22 mm, resistant ≤ 21 mm; for CoNS: susceptible ≥ 25 mm, resistant ≤ 24 mm) and the results of MecA PCR assay.
DNA extraction
Bacterial DNA extraction was performed directly from positive blood cultures. DNA was extracted from positive blood culture fluid as described by Fredricks and Relman [20] using the benzyl alcohol/guanidine hydrochloride organic extraction. To reduce the time required to air-dry the precipitated DNA, we followed the modifications described by Hogg et al. [21] as follows; the DNA pellet was washed and eluted through a Qiagen® spin column, the column was incubated at room temperature for 5 min and centrifuged at 6000 × g for 1 min. The DNA extract was stored at −20 °C.
LAMP assay
In order to distinguish between S. aureus and CoNS and to define oxacillin resistance, the FemA and MecA genes respectively were targeted in the LAMP assay. Primers for the LAMP assay have been published previously [22]. For each target gene, a set of primers to target 8 distinct regions were included to accelerate the LAMP reaction (Table 1) [23].
The LAMP reaction was made in 25 μL by mixing 1.6 μM each inner primer, 0.2 μM each outer primer, 0.8 μM each loop primer, 20 mM tris-HCl, 10 mM KCl , 10 mM (NH4)2SO4, 1.4 mM dNTP, 9 mM MgSO4, 0.8 M Betain (Sigma-Aldrich), 8 U Bst DNA polymerase (New England Biolabs). The mixture was incubated at 63 °C for 45 min and the reaction was heat-stopped at 80 °C for 2 min.
SYBR® Green (Invitrogen) 0.1% was added to each reaction tube. Positive reaction containing LAMP products was visualized with the naked eye as well as by imaging under an ultraviolet (UV) light source (Figure 1); during the initial optimization experiments, amplicons were evaluated by electrophoresis in 2% agarose gel (0.5 × tris-borate-EDTA) (Figure 2). Samples were tested in duplicates to ensure reproducibility.
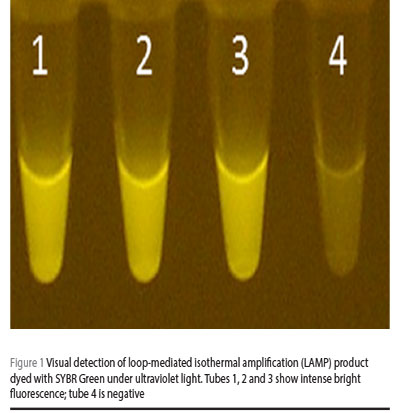
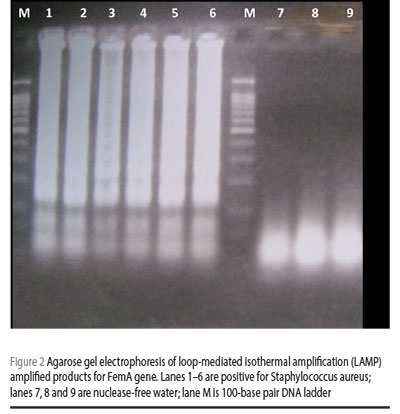
Results
The 60 signal-positive blood culture bottles with growth of GPCC revealed by Gram staining were tested using standard microbiological methods along with MecA PCR as the gold standard. The samples were identified as 19 MRSA, 17 methicillin-sensitive S. aureus (MSSA), 12 methicillin-resistant coagulase-negative staphylococci (MRCoNS) and 9 methicillin-sensitive coagulase-negative staphylococci (MSCoNS); 1 culture grew Micrococcus luteus and 2 cultures yielded no growth on subculture.
LAMP assay
The specimens were tested by the LAMP assay for the presence of the MecA gene encoding oxacillin resistance and for the S. aureus-specific FemA gene. The LAMP successfully amplified the genes under isothermal conditions at 63 °C; all 36 S. aureus isolates were correctly identified when compared with standard laboratory procedures. The identification of MRSA was correctly made for all 19 isolates; the remaining 17 isolates were correctly identified as MSSA (Table 2). Accordingly, for all S. aureus isolates, the method showed 100% sensitivity and specificity and predictive values of 100% for positive or negative test results.
When the LAMP assay was used for detection of the MecA gene in CoNS (FemA negative specimens), 1/12 MRCoNS tested was negative for MecA (shown in bold type in Table 2), giving a sensitivity of 91.7% and negative predictive value 90.0%. The positive predictive value was 100%, with no false positives observed. All blood cultures (n = 15) that showed no growth after 5 days incubation in the BacT/Alert instrument were negative by the LAMP method.
Direct tube coagulase and DCDD test
As shown in Table 3, using standardized phenotypic methods and MecA-PCR, all S. aureus samples that had zone sizes ≤ 19 mm with the DCDD test (n = 15) were confirmed as oxacillin resistant, and all samples with zone sizes ≥ 22 mm with this method (n = 14) were confirmed to be oxacillin susceptible. Thus, accurate and direct detection of oxacillin resistance in S. aureus isolates could be achieved 1 day earlier for 29/36 (80.6%) of blood culture samples, with 100% sensitivity and specificity. Nevertheless, there were 7/36 isolates with zone sizes 20 mm or 21 mm, which represented a mixture of both MRSA and MSSA isolates (shown in bold type in Table 3). Hence, isolates with these zone sizes could not be determined to be oxacillin resistant or susceptible using direct testing, giving an overall sensitivity and specificity of the test of 78.9% and 82.4% respectively.
As regards the use of DCDD in CoNS isolates, 6/21 isolates (28.6%) with cefoxitin zone sizes 22, 23 or 24 mm (shown in bold type in Table 3) could not be confirmed as oxacillin resistant. Of the isolates with zone sizes ≤ 21 and ≥ 25 mm, 15/21 (71.4%) could be determined as oxacillin resistant or susceptible, with 100% sensitivity and specificity.
Discussion
Rapid and accurate detection of MRSA bacteraemia allows rapid adoption of suitable antibiotic therapy and reduces the empirical use of vancomycin [24]. In the present study, in an attempt to determine a practical and simple diagnostic approach for the timely detection of MRSA from positive blood cultures, we evaluated the utility of 2 low-cost methods: the LAMP and DCDD tests. These methods were selected based on the premise that they would increase the speed of reporting results and obviate the need for specialized equipment or highly trained personnel. Instead, most of the costs are in the form of reagents and supplies, which in the case of LAMP is about 80 US cents per sample (including the extraction step) compared with 40 US cents per sample using DCDD.
We tested a total of 75 samples in blood culture bottles by MRSA LAMP, with successful detection of all cases of MRSA and MSSA. Hence, at least for MecA-negative and FemA-positive blood cultures, empirical therapy could be timely adjusted to isoxazolyl penicillins, thus preventing needless use of antibiotics such as vancomycin or linezolid which are associated with additional side-effects, selection of resistant isolates and higher costs.
LAMP analysis directly from the blood culture bottle required a total time of approximately 120 min (60 min for preparation and 60 min for the reaction) in addition to 50 min to perform the extraction. By comparison, conventional laboratory procedures require between 48 and 72 h. The overall test accuracy for detection of MRSA was 100% and predictive values of 100% for positive and negative test results. The results were superior to those presented by previous studies using real-time PCR in blood culture bottles [25,26].
Nevertheless, 1 CoNS (S. epidermidis) isolate was confirmed to be oxacillin-resistant by phenotypic methods, although this isolate was negative by MecA LAMP assay (predictive value 90.0%). It is improbable that a problem in DNA extraction caused this isolate to be negative, since this should also have happened in S. aureus-positive blood cultures. A likely explanation could be the over-expression of beta-lactamases by this isolate, causing a so-called borderline oxacillin resistance S. aureus [27]. Unfortunately, tests to differentiate borderline oxacillin resistance S. aureus from MRSA were not performed in this study.
In the present study, we used the LAMP method to detect MRSA, because of its simple operation, rapid reaction and ease of use [14,15]. One important feature of LAMP assay is the need for just a heat block or a simple water bath in place of a costly thermal cycler, UV gel documentation and transilluminator, which altogether are essential requirements of any conventional PCR. The large amount of DNA produced and the high specificity of the reaction make it possible to identify amplification just by visual examination of fluorescence or turbidity [15].
By combining the LAMP assay with the direct modified organic extraction of DNA, utilizing commercially available nucleic acid spin columns to decrease the time required to air-dry the precipitated DNA, we describe a simple and relatively easy approach to detect MRSA from positive blood cultures.
An additional incubation-dependent method that was evaluated for detection of MRSA directly from positive blood cultures was the DCDD test along with direct tube coagulase. Although oxacillin disk diffusion has been the traditional method for screening for methicillin resistance, the 30 μg cefoxitin disk test was shown to be more efficient in predicting methicillin resistance. Hence the CLSI recommendation that cefoxitin should be preferred over oxacillin for the recognition of MRSA [16].
In the present study we investigated the utility of the DCDD test directly on samples in blood culture bottles. The breakpoints we used were determined by comparing the cefoxitin zone sizes with the results of standardized methods and MecA PCR, and they did not necessarily coincide with the published CLSI guidelines. Since these data were based on small number of isolates, additional studies with a much larger number of clinical samples are required to confirm the preliminary results.
Accurate and direct detection of methicillin resistance in S. aureus isolates was achieved 1 day earlier for 80.6% of blood culture samples, with 100% sensitivity and specificity. Therefore, this test can provide preliminary results while awaiting the results of the standardized antibiotic susceptibility testing. The materials and expertise needed to perform the DCDD test are just a Muller–Hinton plate and a cefoxitin disk, which already a part of any routine diagnostic microbiology laboratory.
There are several limitations to our study. First, the study used a small sample size. However, the available results provide robust pilot data. Secondly, since this was a report from a single institution, just one blood culture system was in operation. Hence, the performance of these methods, predominantly the extraction step, cannot be guaranteed for other systems. Furthermore, both approaches were not validated for the detection of MRSA growth in the anaerobic blood culture bottle, and it would therefore be useful to perform the current evaluation in different blood culture systems comprising both aerobic and anaerobic bottles.
With the benefit of obtaining reliable results in a few hours as opposed to days, both approaches studied here can be adapted for rapid and accurate detection of MRSA form blood culture bottles. The LAMP method exhibited higher diagnostic accuracy within a time frame of 2 to 3 h after the bottle was identified as positive. On the other hand, the DCDD method was more cost-effective and did not require any additional reagents or supplies other than those normally available in a conventional microbiology laboratory. Both methods are therefore relevant for resource-limited settings.
Acknowledgements
Funding: None.
Competing interests: None declared.
References
- Cosgrove SE et al. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infection Control and Hospital Epidemiology, 2005, 26:166–174.
- Blot SI et al. Outcome and attributable mortality in critically Ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Archives of Internal Medicine, 2002, 162(19):2229–2235.
- Lodise TP, McKinnon PS. Clinical and economic impact of methicillin resistance in patients with Staphylococcus aureus bacteremia. Diagnostic Microbiology and Infectious Disease, 2005, 52:113–122.
- Khatib R et al. Impact of initial antibiotic choice and delayed appropriate treatment on the outcome of Staphylococcus aureus bacteremia. European Journal of Clinical Microbiology & Infectious Diseases, 2006, 25:181–185.
- Pape J, Wadlin J, Nachamkin I. Use of BL CHROMagar MRSA medium for identification of methicillin-resistant Staphylococcus aureus directly from blood cultures. Journal of Clinical Microbiology, 2006, 44:2575–2576.
- Kubina M et al. Oxacillin susceptibility testing of staphylococci directly from Bactec Plus blood cultures by the BBL Crystal MRSA ID system. Journal of Clinical Microbiology, 1999, 37:2034–2036.
- Snyder JW, Munier GK, Johnson CL. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from intensive care unit patients. Journal of Clinical Microbiology, 2010, 48:1305–1309.
- Kelley PG et al. Evaluation of the Xpert™ MRSA/SA blood culture assay for the detection of Staphylococcus aureus including strains with reduced vancomycin susceptibility from blood culture specimens. Diagnostic Microbiology and Infectious Disease, 2011, 70:404–407.
- Jukes L et al. Rapid differentiation of Staphylococcus aureus, Staphylococcus epidermidis and other coagulase-negative staphylococci and methicillin susceptibility testing directly from growth-positive blood cultures by multiplex real-time PCR. Journal of Medical Microbiology, 2010, 59:1456–1461.
- Stratidis J, Bia F, Edberg S. Use of real-time polymerase chain reaction for identification of methicillin-resistant Staphylococcus aureus directly from positive blood culture bottles. Diagnostic Microbiology and Infectious Disease, 2007, 58:199–202.
- Abdel Fattah NS, Darwish YW. Antibiogram testing of pediatric skin infections in the era of methicillin-resistant Staphylococci aureus: an Egyptian University Hospital-based study. International Journal of Dermatology, 2012, 51:1441–1447.
- Sobhy N et al. Community-acquired methicillin-resistant Staphylococcus aureus from skin and soft tissue infections (in a sample of Egyptian population): analysis of Mec gene and staphylococcal cassette chromosome. Brazilian Journal of Infectious Diseases, 2012, 16:426–431.
- Saied T et al. Antimicrobial resistance in pathogens causing nosocomial bloodstream infections in university hospitals in Egypt. American Journal of Infection Control, 2011, 39:e61–e65.
- Niemz A, Ferguson TM, Boyle DS. Point-of-care nucleic acid testing for infectious diseases. Trends in Biotechnology, 2011, 29:240–250.
- Njiru ZK. Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Neglected Tropical Diseases, 2012, 6(6):e1572.
- Performance standards for antimicrobial susceptibility testing; informational supplement, 21st ed. Wayne, Pennsylvania, Clinical and Laboratory Standards Institute, 2011 (CLSI document M100-S21).
- Kobayashi N et al. Detection of MecA, FemA, and FemB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiology and Infection, 1994, 113:259–266.
- Qian Q, Eichelberger K, Kirby JE. Rapid identification of Staphylococcus aureus in blood cultures by use of the direct tube coagulase test. Journal of Clinical Microbiology, 2007, 45:2267–2269.
- Bennett K, Sharp SE. Rapid differentiation of methicillin-resistant Staphylococcus aureus and methicillin-susceptible Staphylococcus aureus from blood cultures by use of a direct cefoxitin disk diffusion test. Journal of Clinical Microbiology, 2008, 46:3836–3838.
- Fredricks DN, Relman DA. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. Journal of Clinical Microbiology, 1998, 36:2810–2816.
- Hogg GM, McKenna JP, Ong G. Rapid detection of methicillin-susceptible and methicillin-resistant Staphylococcus aureus directly from positive BacT/Alert blood culture bottles using real-time polymerase chain reaction: evaluation and comparison of 4 DNA extraction methods. Diagnostic Microbiology and Infectious Disease, 2008, 61:446–452.
- Xu Z et al. Development and application of loop-mediated isothermal amplification assays on rapid detection of various types of staphylococci strains. Food Research International, 2012, 47(2):166–173.
- Notomi T et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research, 2000, 28(12):e63.
- Shime N, Kosaka T, Fujita N. The importance of a judicious and early empiric choice of antimicrobial for methicillin-resistant Staphylococcus aureus bacteraemia. European Journal of Clinical Microbiology and Infectious Diseases, 2010, 29:1475–1479.
- Tan TY et al. Rapid identification of methicillin-resistant Staphylococcus aureus from positive blood cultures by real-time fluorescence PCR. Journal of Clinical Microbiology, 2001, 39:4529–4531.
- Thomas LC et al. Development of a real-time Staphylococcus aureus and MRSA (SAM-) PCR for routine blood culture. Journal of Microbiological Methods, 2007, 68:296–302.
- Keseru JS et al. Investigation of beta-Lactamases in clinical isolates of Staphylococcus aureus for further explanation of borderline methicillin resistance. Chemotherapy, 2005, 51:300–304.


