Research article
E. Hagag,1,2 M.Shwaireb,1 J. Coffa 3 and A. El Wakil 1,2
تحري إعادة الترتيب الجينومي الواسع في الجين BRCA1 لدى المصابات بسرطان الثدي الوراثي
إيمان حجاج، محمد شويرب، جوردي كوفا، عبير الوكيل
الخلاصـة: ينتقل ما يقرب من %5 إلى %10 من سرطانات الثدي وراثياً نتيجة طفرات في الخلايا الجنسية في الجين BRCA1. ولم تتم دراسات بحثية على إعادة الترتيب الواسع في هذا الجين لدى المصريات. وقد استخدم الباحثون التضخيم المتعدد الذي يعتمد على الربط بالمسابر لتوضيح إعادة ترتيب الجين BRCA1 لدى 4 حالات من بين 22 حالة (18.2%) من حالات سرطان الثدي العائلي. ولم يلاحظوا أي تأثير لوجود حالات متعددة من سرطان الثدي في العائلة لدى المريضات اللاتي تم تشخيصهن بين 30 و45 عاماً ولديهن إيجابية في إعادة الترتيب الواسع في الجين BRCA1. إلا أنه عند التركيز على الحالات التي كانت الإصابة فيها في الأقارب من الدرجة الأولى أو الثانية، لاحظ الباحثون اختلافاً هاماً في إعادة الترتيب الواسع في الجينوم بين نسبة المريضات الإيجابيات لجين BRCA1 مقابل السلبيات لجين BRCA1. وتقدم النتائج التي توصل إليها الباحثون أولى البيِّنات على وجود إعادة الترتيب الواسع في الجينوم في الجين BRCA1 لدى المصريات. وقد يكون من المرغوب تحري هذه الطفرات عند تشخيص سرطان الثدي في عمر مبكر، ولاسيَّما في الحالات التي تكون هناك إصابة مماثلة لدى أقارب من الدرجة الأولى أو الثانية.
ABSTRACT Approximately 5%–10% of all breast cancers are inherited as the result of germline mutations in the BRCA1 gene. Large genomic rearrangements (LGRs) in BRCA1 have not been well-researched in the Egyptian population. Using multiplex ligation-dependent probe amplification, we showed BRCA1 rearrangements in 4/22 cases (18.2%) of familial breast cancer. No influence of having multiple breast cancer cases within the family was observed in patients diagnosed at < or ≥ 45 years and having BRCA1-positive LGRs. However, focusing on cases with first- and second-degree relatives affected, we observed a significant difference between the percentage of patients with BRCA1-positive versus BRCA1-negative LGRs. Our results provide the first evidence that LGRs in BRCA1 exist in the Egyptian population. Screening for these alterations may be desirable when breast cancer patients are diagnosed at an early age, especially if these cases have first- and second-degree of relatives with breast cancer.
Dépistage de grands réarrangements génomiques sur le BRCA1 chez des patientes égyptiennes atteintes d'un cancer du sein héréditaire
RÉSUMÉ Des mutations germinales sur le gène BRCA1 sont responsables d'environ 5 à 10 % de tous les cancers du sein. Les grands réarrangements génomiques sur le gène BRCA1 n'ont pas été bien étudiés dans la population égyptienne. L'amplification multiplex de sonde nucléique dépendant des ligatures nous a permis de mettre en évidence des réarrangements sur le gène BRCA1 dans 4 cas sur 22 (18,2 %) de cancer du sein familial. Le fait d'avoir plusieurs cas de cancer du sein dans une même famille n'avait pas d'influence chez les patientes ayant reçu le diagnostic à un âge inférieur, égal ou supérieur à 45 ans et présentant de grands réarrangements génomiques du gène BRCA1. Toutefois, après une étude plus approfondie des cas ayant des parentes au premier et second degré touchées, nous avons observé une différence significative entre le pourcentage de patientes positives pour les grands réarrangements génomiques sur le gène BRCA1 et celui des patientes dont les analyses étaient négatives en la matière. Nos résultats fournissent la première preuve selon laquelle les grands réarrangements génomiques sur le gène BRCA1 existent dans la population égyptienne. Le dépistage de ces altérations peut être souhaitable lorsque les patientes atteintes du cancer du sein reçoivent le diagnostic à un âge assez jeune, en particulier lorsque ces cas ont des parentes au premier et deuxième degré également atteintes d'un cancer du sein.
1Department of Biological and Geological Sciences, Faculty of Education, University of Alexandria, Alexandria, Egypt.
2Institute of Molecular and Cellular Pharmacology, University of Nice–Sophia Antipolis, Nice, France (Correspondence to A. El Wakil::
3MRC–Holland b.v., Amsterdam, The Netherlands.
Received: 18/12/11; accepted: 06/03/12
EMHJ, 2013, 19(3):255-262
Introduction
Breast cancer is one of the most common cancers affecting women worldwide [1,2]. In Egypt, it ranks number one among female malignancies, representing 18.9% of total cancer cases in the year 2001 [3] and is responsible for 15% of cancer deaths in Egyptian women [4]. Although epidemiological investigations have identified numerous risk factors for breast cancer, a positive family history or genetic factor has been identified as a major contributor to the risk of developing this disease. Confirmation of this hypothesis was achieved by the discovery of the breast cancer susceptibility gene, BRCA1 (GenBank accession no. U14680, OMIM 113705). Approximately 5%–10% of all breast cancers are inherited as the result of germline mutations in this gene in an autosomal dominant fashion [5–7]. Women with germline mutations in BRCA1 have a lifetime risk of 85% for breast cancer [8,9]
The demand for screening for BRCA1 mutations is increasing, as their identification will affect the medical management of cases. Traditionally, detection of these types of mutations is done using Southern blot hybridization or fluorescent in situ hybridization techniques, which can be laborious, time-consuming, and require high quantities of starting material [3]. The multiplex ligation-dependent probe amplification (MLPA) method has emerged in recent years as a powerful and reliable method to screen for large genomic rearrangements (LGRs) in BRCA1, among others [10–13].
It is now clear that, as with small mutations, the contribution as well as the diversity of rearrangement-associated mutations of LGRs is variable between different populations. Indeed, the incidence of BRCA1 LGRs in patients with strong family history of breast cancer ranges from 0.8%–23%, which represents around 8%–40% contribution to the BRCA1 mutation spectrum in different ethnic groups [14–17]. LGRs in the BRCA1 gene in Egyptian population have not been widely researched [3]. Therefore, a comprehensive analysis of these rearrangements in an Egyptian patient cohort was needed.
The main objective of the present work was to identify LGRs in the BRCA1 gene using the MLPA technique in a cohort of 22 high-risk Egyptian female patients. To the best of our knowledge, this is the first study using MLPA to search for LGRs in the Egyptian population. The information aimed to improve our understanding of the nature and frequency of these genetic events in Egypt.
Methods
Patients and samples
A total of 22 Egyptian high-risk breast cancer female patients were enrolled from the Medical Research Institute, University of Alexandria, after they had given informed consent. Our study protocol was approved by the ethics committee of the Institute. Patients were characterized according to clinicopathological features and family history, as shown in Table 1. Eligible for the current study were patients classified as having an increased risk of carrying BRCA mutations due to their family history. They were categorized into the following risk groups:
Group A: 11 patients having 1 or more cases of breast cancer in first- or second-degree relatives (2 patients had 2 other female relatives affected by the same cancer type).
Group B: 9 patients having 1 case of breast cancer in third- or fourth-degree relatives (2 patients were diagnosed ≤ age 30 years).
Group C: 2 patients having at least 3 cases of breast cancer in third- or fourth-degree relatives.
Late presentation is a characteristic feature of cases in Egypt. All patients were suffering from invasive duct carcinoma. Pathological grading showed no incidence of grade I, whereas the incidence of grade II and III tumours was 77.3% and 22.7% respectively. Breast cancer in Egyptian women is typically detected at a young age and the majority of our cases were 35–55 years of age (Table 1); the mean age was 45 years. Most patients were premenopausal (17/22 patients, 77.3%). The mean tumour size was 3.5 cm. The frequency of positive axillary lymph node metastases was 77.3% (17/22 patients). The profile of hormone receptors as determined by immunohistochemistry was positive for oestrogen receptors in 81.8%, for progesterone receptors in 77.3% and for both receptors in 63.6% of cases.
MLPA assay
All screened women provided a 10 mL blood sample in addition to DNA samples obtained from 4 healthy volunteers to serve as controls. Genomic DNA was extracted from whole blood samples using the AxyPrep Blood Genomic DNA Miniprep kit (Axygen Biosciences). MLPA technique was performed on genomic DNA to detect BRCA1 LGRs, using the SALSA MLPA kits for BRCA1, primary screening kit P002-C1 and confirmation kit P087 (MRC–Holland b.v.) according to the manufacturer’s instructions. Fragment analysis was carried out on ABI-3100 XL genetic analyser (Applied Biosystems), using LIZ-500 (Applied Biosystems) as the DNA molecular marker and standard electrophoretic conditions. When a positive result (40% change) appeared, the analysis was confirmed by the MLPA confirmation kit P087.
Data analysis
Following fragment separation by capillary gel electrophoresis, the obtained PCR product data were quantified and interpreted using the MLPA analysis software Coffalyser.NET. In short, the unique length of every probe in the MLPA probe mix is used to associate the detected signals to the original probe sequences. These probe product measurements are proportional to the amount of the target sequences present in a sample. In order to make these data intelligible, unknown samples need to be compared with reference samples, which can be done by normalization. After normalization the relative amount of fluorescence related to each probe can be expressed in dosage quotients, which is the usual method of interpreting MLPA data. This dosage quotient or ratio is a measure for the ratio in which the target sequence is present in the sample DNA as compared to the reference DNA, or relative ploidy. During normalization our software made use of every reference probe for normalization of each test probe, thereby producing as many dosage quotients as there are references probes. The median of these dosage quotients was then be used as the definite ratio.
Statistical analysis
The data were analysed using StatPac statistics calculator software. Statistical differences were determined with Fisher exact test or the 2 sample t-test between percentages. The criterion for significant difference was set at P ≤ 0.05.
Results
Using MLPA analysis, we identified LGRs in 4 out of 22 high-risk female breast cancer patients (18.2%). Figure 1 shows the electropherograms in a control sample (Figure 1a) and patient samples (Figures 1b–d). Apart from recurrent aberrations, such as BRCA1 exon 1a duplication in 1 patient (4.5%) (Figure 1c) and exon 22 duplication in 1 patient (4.5%) (Figure 1d), our data could fully characterize 1 novel LGR in these patients (Figure 1b). This alteration identified the duplication of exons 3, 22 and 24 in 2 patients (9.1%). All BRCA1 rearrangements were verified and confirmed in a new DNA sample using the P087 BRCA1 MLPA control kit.
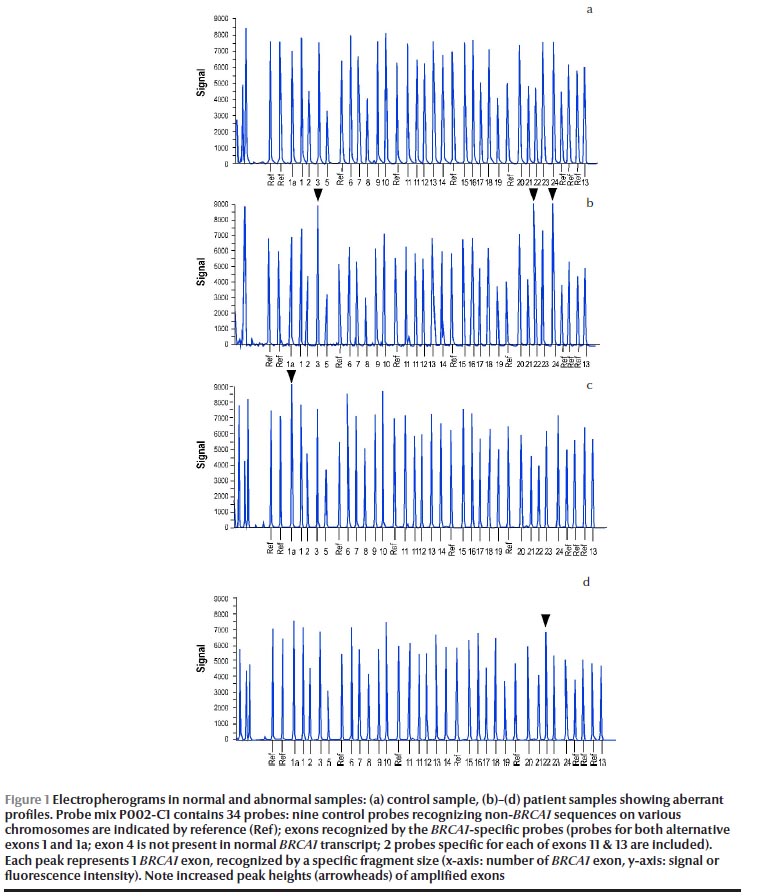
This comprehensive analysis of the BRCA1 mutation status category showed the highest positive LGRs frequency in patients of group B (3/9, 33.3%,), whereas the mutation frequencies were clearly lower in group A (1/9, 9.1%) and no mutations were reported in group C.
Focusing on group B as it had the highest frequency of LGRs, the mean age of onset of breast cancer was 46.3 years, whereas the mean age for patients harbouring LGRs was 38.6 years. However, our data showed no significant difference between the pathogenic BRCA1 mutation status category in patients before or after the mean age of 45 years (P = 0.12, Fisher exact test). In the 2 cases in which breast cancer was diagnosed at age 30 years, 1 was BRCA1-positive and 1 was BRCA1-negative for LGRs.
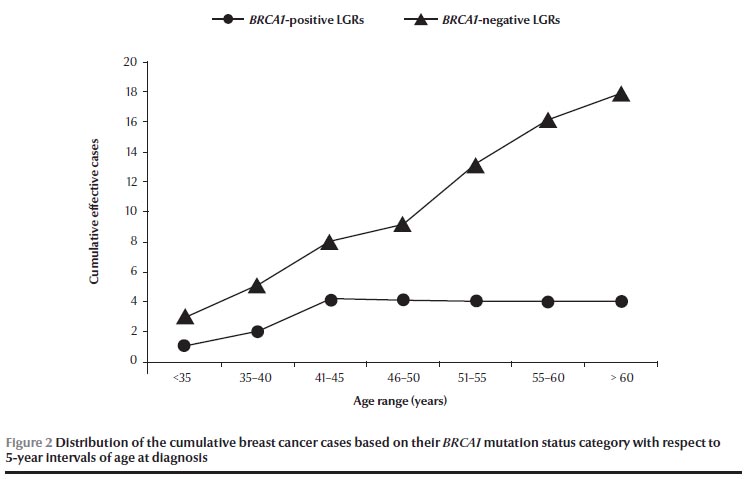
Analysing the total 22 cancer patients, a phenotypic characterization based on pathogenic mutation status revealed that the age at diagnosis was younger (≤ 45 years) in patients with BRCA1-positive (4/4, 100%) than in patients with BRCA1-negative LGRs (8/18, 44.4%). Consequently, the distribution of the cumulative cases of breast cancer increased over 5-year intervals only in BRCA1-negative LGRs (Figure 2). However, no significant difference was found in the BRCA1 mutation status category of patients either younger or older than the mean age of 45 years old (P = 0.07, Fisher exact test).
We also investigated the relationship between the number of cases of breast cancer within the family of each patient (≤ 2 or ≥ 3 cases) based on the pathogenic BRCA1 mutation status category and age at diagnosis < or ≥ 45 years (Table 2 ). Among these 2 groups of patients having either ≤ 2 or ≥ 3 breast cancer cases within their family history, no significant difference was found either between the pathogenic BRCA1 mutation status category (P = 0.93, Fisher exact test) or between the age of patients at diagnosis (P = 1.00, Fisher exact test). Moreover, among the BRCA1- positive LGRs patients, no significant difference between the percentage of breast cancer cases diagnosed at < 45 years of age was found within groups having either ≥ 3 cases (50%, 1 patient out of 2) or those having ≤ 2 cases (30%, 3 patients out of 10) of breast cancer within their family history in a comparative analysis of 2 sample t-test between percents (t = 0.548, df = 10, P = 0.59).
The analysis was extended to evaluate the BRCA1 mutation status, comparing cases with first- or second-relatives and third- or fourth-degree relatives based on the average age of 45 years (Table 3 ). Among the BRCA1-positive patients there were no significant differences in the distribution of cases either comparing mutation-negative versus mutation-positive groups (P = 0.29), or comparing cases younger versus older than the mean age of 45 years (P = 1.00). This can be attributed to the small number of patients in the cohort studied. However, focusing only on the first- or second-degree relatives group, we observed a significant difference between the percentage of patients with BRCA1-positive versus BRCA1-negative LGRs in a 2-sample t-test comparison between percentages (t = 3.846, df = 20, P = 0.001).
Discussion
The presence of genomic rearrangements of the BRCA1 gene in breast cancer have been intensively investigated in patients from various countries over recent years. A number of different rearrangements have been reported that clearly document the involvement of this mutation type in genetic predisposition to breast cancer [18]. Population-specific studies are now needed to evaluate the prevalence of genomic rearrangements before deciding whether to include ad hoc screening procedures into standard diagnostic mutation detection strategies. In Egypt, other studies have analysed the BRCA1 mutations; however, all were based on PCR-based screening methods [3] and consequently, no LGRs in the BRCA1 locus have been reported so far in Egyptian women with a positive family history of breast cancer. Here we report the results of the first quantitative genomic analysis by the MLPA technique of the BRCA1 gene in Egyptian high-risk breast cancer female patients in whom previous analyses of the BRCA1 gene by conventional scanning methods failed to identify mutations.
Genomic rearrangements accounted for 18.2% (4/22) of the BRCA1 mutations detected in these patients. This rate is somewhat higher than those previously recorded in other populations such as in Spain (8.2%) [19], Germany (9.6%) [20], France (12.0%) [5], Czech Republic (12.3%) [21] and Denmark (12.5%) [22], but is lower than that recorded in the Netherlands (27.3%) [13]. Our reported percentage is in agreement with the results of Agata et al. in Italy, who reported about 19% of LGRs in a large cohort of breast and breast/ovarian cancer families without BRCA1 and BRCA2 point mutations detectable by conventional scanning methods [14].
Age is an important risk factor for breast cancer. The rate of positive BRCA1 mutations in patients at an early age at diagnosis (≤ 30 years) and with negative family history was found to be 12.9% in Czech patients [23] and 2% in English women [24]. So we could predict that the rate would be much higher in patients diagnosed at an earlier age as well as those with a positive family history. This is consistent with our data on the occurrence of BRCA1-positive mutations in 33.3% (1/3) of patients diagnosed at ≤ 30 years old. Furthermore, in accordance with this, we report a higher percentage of younger patients at diagnosis in the BRCA1-positive category (100%, 4/4) than in the BRCA1-negative category (44.4%, 8/18). So the high correlation between age and genetic risk attributable to BRCA1 could reflect the influence of family history [25].
Based on studies of age at diagnosis another research group concluded that the BRCA1 mutation was a strong candidate for screening for early onset breast cancer [26]. Our data revealed that BRCA1-positive women were on average about 10 years younger (38.6 years) than BRCA1-mutation negative women (48.4 years). Consequently, we found 33.3% (4/12) of patients diagnosed at ≤ 45 years had BRCA1-positive LGRs and 33.3% (1/3) diagnosed at ≤ 30 years harboured BRCA1-positive LGRs. Furthermore, to be able to compare our results with those of Ibrahim et al.’s recent study of on mutations in an Egyptian population, we limited the age at onset of diagnosis to 40 years and not 45 years old [3]. In the present study, 2 patients (9.1%) were diagnosed as BRCA1-positive at ≤ 40 years old, which is lower than the percentage reported by Ibrahim et al. [3], although the patients in their study had a negative family history unlike the patients in our study who all had a positive family history.
Furthermore, younger age at diagnosis and the occurrence of multiple cases of breast cancer within the family history were previously reported to be the strongest predictors of the likelihood of carrying a BRCA1 mutation [27]. This is not in agreement with our results, as we reported no significant correlation between patients diagnosed at ≤ 45 years of age and having ≥ 3 cases of breast cancer within their family history and the BRCA1-positive mutation status.
One limitation of our study was the small number of patients tested. Egypt is known for a considerable degree of inbreeding. This makes it necessary to screen a large number of patients in order to get a true picture of the contribution of BRCA1 to familial breast cancer. However, this limited study with 22 high-risk patients suggested that BRCA1 mutation does play a key role in the development of familial breast cancer among Egyptian women as 18.2% of patients screened for the presence of LGRs in BRCA1 were found to be positive.
In summary, the present study showed that BRCA1 rearrangements accounted for a relatively large proportion of familial breast cancer cases in Egyptian population. Nevertheless, no reliable data about the spectrum and precise prevalence of specific gross rearrangements were reported due to the low number of patients recruited. As far as we know, our results provide the first evidence that, as in many other studied populations, large genomic changes in BRCA1 do also exist in Egypt. Screening for these alterations as part of comprehensive genetic testing in the Egyptian hereditary breast cancer patients may, therefore, be recommended in patients diagnosed at an early age, especially if these cases have first- and second-degree of relatives within their family history.
Acknowledgements
We are grateful to Dr Eman El Abd, Molecular Biology Group, Medical Technology center, Medical Research Institute, University of Alexandria, Egypt, for locally providing the samples and their related diagnosis needed for this study. We thank Nathalie Leroudier and Nicole Zsürger for technical assistance, and Franck Aguila for art work. We wish also to thank Dr Enzo Lalli for helpful discussions and critical reading of the manuscript. This study was supported by a mobility grant offered by the AUF (Agence Universitaire de la Francophonie) to the master student, Eman Hagag.
The study was funded by the Agence Universitaire de la Francophonie (AUF), Bureau Moyen Orient, which offered a mobility grant to the master student and the MRC–Holland, Netherlands, provided additional MLPA kits.
References
- Jemal A et al. Cancer statistics, 2008. CA: a Cancer Journal for Clinicians, 2008, 58:71–96.
- Boulos S et al. Breast screening in the emerging world: high prevalence of breast cancer in Cairo. Breast, 2005, 14:340–346.
- Ibrahim SS, Hafez EE, Hashishe MM. Presymptomatic breast cancer in Egypt: role of BRCA1 and BRCA2 tumor suppressor genes mutations detection. Journal of Experimental and Clinical Cancer Research, 2010, 29:82.
- Omar S et al. Breast cancer in Egypt: a review of disease presentation and detection strategies. Eastern Mediterranean Health Journal, 2003, 9:448–463.
- Gad S et al. Significant contribution of large BRCA1 gene rearrangements in 120 French breast and ovarian cancer families. Oncogene, 2002, 21:6841–6847.
- Preisler-Adams S et al. Gross rearrangements in BRCA1 but not BRCA2 play a notable role in predisposition to breast and ovarian cancer in high-risk families of German origin. Cancer Genetics and Cytogenetics, 2006, 168:44–49.
- Cavallone L et al. Comprehensive BRCA1 and BRCA2 mutation analyses and review of French Canadian families with at least three cases of breast cancer. Familial Cancer, 2010, 9:507–517.
- Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Research and Treatment, 2011, 125:325–349.
- del Valle J et al. Identification and comprehensive characterization of large genomic rearrangements in the BRCA1 and BRCA2 genes. Breast Cancer Research and Treatment, 2010, 122:733–743.
- Schouten JP et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Research, 2002, 30:e57.
- Jankowski S et al. Multiplex ligation-dependent probe amplification analysis on capillary electrophoresis instruments for a rapid gene copy number study. Journal of Biomolecular Techniques, 2008, 19:238–243.
- Coffa J et al. MLPAnalyzer: data analysis tool for reliable automated normalization of MLPA fragment data. Cellular Oncology, 2008, 30:323–335.
- Hogervorst FB et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Research, 2003, 63:1449–1453.
- Agata S et al. Prevalence of BRCA1 genomic rearrangements in a large cohort of Italian breast and breast/ovarian cancer families without detectable BRCA1 and BRCA2 point mutations. Genes, Chromosomes & Cancer, 2006, 45:791–797.
- Peixoto A et al. BRCA1 and BRCA2 germline mutational spectrum and evidence for genetic anticipation in Portuguese breast/ovarian cancer families. Familial Cancer, 2006, 5:379–387.
- Vasickova P et al. High occurrence of BRCA1 intragenic rearrangements in hereditary breast and ovarian cancer syndrome in the Czech Republic. BMC Medical Genetics, 2007, 8:32.
- Pylkäs K et al. Analysis of large deletions in BRCA1, BRCA2 and PALB2 genes in Finnish breast and ovarian cancer families. BMC Cancer, 2008, 8:146.
- Ford D et al.; The Breast Cancer Linkage Consortium. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. American Journal of Human Genetics, 1998, 62:676–689
- de la Hoya M et al. Genomic rearrangements at the BRCA1 locus in Spanish families with breast/ovarian cancer. Clinical Chemistry, 2006, 52:1480–1485.
- Engert S et al. MLPA screening in the BRCA1 gene from 1,506 German hereditary breast cancer cases: novel deletions, frequent involvement of exon 17, and occurrence in single early-onset cases. Human Mutation, 2008, 29:948–958.
- Ticha I et al. Screening for genomic rearrangements in BRCA1 and BRCA2 genes in Czech high-risk breast/ovarian cancer patients: high proportion of population specific alterations in BRCA1 gene. Breast Cancer Research and Treatment, 2010, 124:337–347.
- Hansen T et al. Large BRCA1 and BRCA2 genomic rearrangements in Danish high risk breast-ovarian cancer families. Breast Cancer Research and Treatment, 2009, 115:315–323.
- Pohlreich P et al. High proportion of recurrent germline mutations in the BRCA1 gene in breast and ovarian cancer patients from the Prague area. Breast Cancer Research, 2005, 7:R728–R736.
- Lalloo F et al.; Early Onset Breast Cancer Study Group. Prediction of pathogenic mutations in patients with early-onset breast cancer by family history. Lancet, 2003, 361:1101–1102.
- Díez O et al. BRCA1 mutation analysis in 83 Spanish breast and breast/ovarian cancer families. International Journal of Cancer, 1999, 83:465–469.
- Aktas D et al. Identification of point mutations and large rearrangements in the BRCA1 gene in 667 Turkish unselected ovarian cancer patients. Gynecologic Oncology, 2010, 119:131–135.
- Woodward AM et al.kConFab Investigators. Large genomic rearrangements of both BRCA2 and BRCA1 are a feature of the inherited breast/ovarian cancer phenotype in selected families. Journal of Medical Genetics, 2005, 42:e31.


