Messis Abdelaziz1,2, Adjebli Ahmed3, Ayeche Riad4, Ghidouche Abderrezak2; Ait-Ali Djida2
1Université de Bordj Bou Arréridj, El-Anasser, 34010, Bordj Bou Arréridj, Algérie; 2Laboratoire de Génie Biologique des Cancers, Université de Bejaia 06000, Bejaia, Algérie; 3Laboratoire d’Ecologie Microbienne, faculté des sciences de la nature et de la vie, université de Bejaia, 06000, Bejaia, Algérie; 4Laboratoire Caractérisation et Valorisation des Ressources Naturelles, Université de Bordj Bou Arreridj, 34010, El-Anasser, Bordj Bou Arréridj, Algérie. (Correspondence to:
ABSTRACT
Background: COVID-19 has become a worldwide threat affecting every country.
Aims: This study aimed to identify COVID-19 cases in Algeria using times series models for forecasting COVID-19.
Methods: Confirmed COVID-19 daily cases data were obtained from 21 March 2020 to 26 November 2020 from the Algerian Ministry of Health. Forecasting was done using the Autoregressive Integrated Moving Average (ARIMA) models (0,1,1) with Minitab 17 software.
Results: Observed cases during the forecast period were accurately predicted and placed within prediction intervals generated by ARIMA. Forecasted values of COVID-19 positives, recoveries and deaths showed an accurate trend, which corresponded to actual cases reported during 252, 253 and 254 days. Results were strengthened by variations of less than 5% between forecast and observed cases in 100% of forecasted data.
Conclusion: ARIMA models with optimally selected covariates are useful tools for predicting COVID-19 cases in Algeria.
Keywords: COVID-19, time series, double exponential smoothing, ARIMA, forecast, Algeria
Citation: Abdelaziz M, Ahmed A, Riad A, Abderrezak G2, Djida Ait-Ali. Forecasting daily confirmed COVID-19 cases in Algeria using ARIMA models. East Mediterr Health J. 2023;29(5):xxx–xxx. https://doi.org/10.26719/emhj.23.054
Received: 13/03/21; accepted: 08/12/22
Copyright: ©Authors; Licensee: World Health Organization. EMHJ is an open access journal. All papers published in EMHJ are available under the Creative Commons Attribution Non-Commercial ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo).
Introduction
On 11 March 2020, the World Health Organization (WHO) declared COVID-19 as a pandemic. The pandemic had spread from mainland China to other countries and territories, disrupting socioeconomic activities. As of 26 November 2020, COVID-19 had infected more than 60 776 978 people globally, killed more than 1 428 228, and resulted in a lockdown that forced people to stay in their homes (1).
Algeria reported its first COVID-19 case on 25 February 2020. By 26 November, it had reported 79 110 confirmed cases, 51 334 recoveries and 2352 deaths (2).
SARS-CoV-2, the COVID-19 virus is very infectious, and many people were not following the non-pharmaceutical public health prevention measures recommended by the Algerian government and other governments to control the pandemic, thus increasing the risks of transmission (2,3).
Accurate forecasting of COVID-19 case trends was essential for preparedness by health authorities to manage the pandemic and resource planning. Time series models such as ARIMA have been widely used to statistically model and forecast infectious disease trends (4). ARIMA models are preferred in this context because they are suitable for investigating short-term effects of acute infectious diseases and are flexible and appropriate for several trajectories (4,5). ARIMA models have been used in several studies to forecast COVID-19 outbreak trends (6-9).
In this study, we developed ARIMA models using daily COVID-19 confirmed and active cases in Algeria to identify the best fitting model of COVID-19 cases from 21 March 2020 to 26 November 2020.
Materials and methods
Data source
Data for this study included confirmed COVID-19 daily cases data obtained from 21 March 2020 to 26 November 2020 from the Algerian Ministry of Health (10).
Methods
The following equation highlights the exponential smoothing method and the ARIMA processes (11):

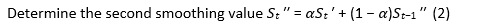
ARIMA model for time series sata ARIMA model is stated as follows:

Where:
ɸ(B) is an autoregressive operator
Ө(B) is a moving average operator
(1 − B)d is a differencing operator. It is the expression of dth consecutive differencing so as to make the series stationary
Zt is a Gaussian white noise series with mean zero and variance (σ2z).
ARIMA forecast is based on previous values and portrayed by 3 terms – p, d, q. Where p is the order for the auto regressive expression (AR), q is the order for the moving average expression (MA) and d is the number of differencing required making the time arrangement fixed.
The experiment was carried out using Minitab 17 programming software (12). In general, the equation can be approached using a regression model:
εt = errors from the accompanying conditions.
Results
Using the time-series model approach, the pattern of COVID-19 data distribution in Algeria showed an exponential distribution pattern, where the addition of positive cases increased significantly everyday of the pandemic. The distribution pattern was the same for the number of people who recovered and died (Figure 1). For the positive COVID-19 cases, the mean absolute percentage error (MAPE) value was smaller than the error rate at 10% (Table 1). The increase in the number of people who were positive for COVID-19 directly affected the prediction model for patients who recovered and died (Figure 1). For recovered cases, MAPE value was smaller than the error value set at 10% error rate (Table 1). The recovery rate for COVID-19 patients increased simultaneously with the number of positive cases because of the non-pharmaceutical public health measures taken by the government from 21 March 2020. For deaths due to COVID- 19, the MAPE value was greater than the error value set at 10% (Table 1). The increase in mortality was possibly due to the extent of infection and the medical history of the patients.
In the time series model with 5% error probability (α), the graph followed the ARIMA process (0,1,1), with the P value MA 1 (0.0%) smaller than α.
Estimated results of parameters model for COVID-19 positive data using ARIMA models:
Referring to equation (4), mathematically, the ARIMA model (0,1,1) can be stated using the following coefficients:
γt = 317.65 − 0.879et-1
Same as COVID-19 positive data, in the time series model with 5% error probability (α), the graph followed the ARIMA process (0,1,1) with the P value MA 1 (0,0%) smaller than α.
Estimated of parameters for COVID-19 Recovery data results using ARIMA model:
Referring to equation (4), mathematically the ARIMA model (0,1,1) can be stated as follows: γt = 205.53 − 0.30et-1
After the positive and recovery data were analysed in time series model with the 5% error probability (α), the graph followed the ARIMA process (0,1,1) with the P value MA 1 (0.00%) smaller than α.
All estimated parameter results of the ARIMA model:
Referring to equation (4), mathematically, the ARIMA model (0,1,1) can be stated.
The results of predictions of COVID-19 cases in Algeria (positive, recovered and deaths) showed a gap in the resulting distribution patterns, where the increase in the number of positive cases was not offset by an increase in the number of patients who recovered and a decrease in the number of patients who died. This indicates that public behaviour did not comply with the rules set by the government (physical distancing, large-scale social restrictions, washing of hands, and mask use).
Discussion
From the time WHO declared COVID-19 a pandemic on 11 March 2020, several countries experienced an exponential increase in COVID-19 cases (3), which put a lot of pressure on most healthcare systems worldwide. In response, health authorities attempted to forecast the trend of the pandemic, but this proved difficult because COVID-19 is a novel disease with limited data and knowledge about its trends and dynamics (2). Our forecast showed an accurate trend, which corresponded to the number of positive cases observed and reported by the Ministry of Health in Algeria during three days (252, 253 and 254). The same situation was observed for forecasted recoveries and deaths.
This finding was strengthened by variations of less than 5% between the forecast and observed cases in 100% of the forecasted data points (Table 2). Similar studies conducted in South Korea, Iran and Italy predicted similar case trends using ARIMA models (6-8).
The strengths of this study include: firstly, this is the first paper to report the use of ARIMA models to forecast COVID-19 cases and trends in Algeria. Secondly, this was the first attempt to use smoothen case data to improve accuracy as compared to similar studies on ARIMA models for COVID-19 conducted in other countries (6-7). Thirdly, we used several independent covariates, which provided more accurate signals to develop short-term model predictions for immediate outbreak response. Finally, we optimized the model training and validation period to provide the highest number of data points to generate the best fit model.
Conclusion
This study demonstrates the effectiveness of ARIMA models as an early warning strategy that can provide accurate COVID-19 forecasts on larger data points (251 days). Forecasted values of COVID-19 positives, recoveries and deaths showed an accurate trend which corresponded to the actual cases observed and reported by the Ministry of Health in Algeria during three days (252, 253 and 254). We are confident that the ARIMA model can be used to generate accurate and reliable forecasts of daily COVID-19 cases until the end of COVID-19, with the addition of new data points and independent covariates.
Declaration of competing interest
I have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors would like to thank the Ministry of Health, Population and Hospital Reform of Algeria and the Johns Hopkins University for publicly releasing the updated COVID-19 datasets.
References
- World Health Organization. Coronavirus disease (COVID-19) outbreak situation. Geneva: World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- Johns Hopkins University. Novel coronavirus (COVID-19) cases. Baltimore: Johns Hopkins University. https://coronavirus.jhu.edu/region/algeria.
- Lounis M. Epdemiology of coronavirus disease 2020 (COVID-19) in Algeria. New Microbes and New Infections 2021;39:100822. DOI: https://doi.org/10.1016/j.nmni.2020.100822.
- Allard R. Use of time-series analysis in infectious disease surveillance. Bull World Health Organ 1998;76:327–333.
- Imai C, Hashizume M. A systematic review of methodology: Time series regression analysis for environmental factors and infectious diseases. Trop Med Health 2015;43:1–9. DOI: https://doi.org/10.2149/tmh.2014-21.
- Benvenuto D, Giovanetti M, Vassallo L, Angeletti S, Ciccozzi M. Application of the ARIMA model on the COVID-2019 epidemic dataset. Data in Brief 2020;29:1-4. DOI: https://doi.org/10.1016/j.dib.2020.105340.
- Chintalapudi N, Battineni G, Amenta F. COVID-19 virus outbreak forecasting of registered and recovered cases after sixty-day lockdown in Italy: A data driven model approach. J Microbiol Immunol Infect 2020;53:396-403. DOI: https://doi.org/10.1016/j.jmii.2020.04.004.
- Singh S, Sundram BM, Rajendran K, Law KB, Aris T, Ibrahim H, Dass SC, Gill BS. Forecasting daily confirmed COVID-19 cases in Malaysia using ARIMA models. J Infect Dev Ctries 2020;14. DOI: https://doi.org/10.3855/jidc.13116.
- Singh RK, Rani M, Bhagavathula AS, Sah R, Rodriguez-Morales AJ, Kalita H, Nanda C, Sharma S, Sharma YD, Rabaan AA, Rahmani J, Kumar P. Prediction of the COVID-19 pandemic for the top 15 affected countries: Advanced Autoregressive Integrated Moving Average (ARIMA) model. JMIR Public Health Surveill 2020;6. DOI: https://doi.org/10.2196/19115.
- Algerian health and hospital reform minister: carte épidémiologique. https://www.covid19.gov.dz/carte/).
- Konarasinghe KMUB. Modeling COVID-19 epidemic of USA, UK, and Russia. JNFHBS 2020;1:1-14.
- Minitab 17 Statistical Software. Minitab, Inc.: State College, PA, USA, 2010. www.minitab.com.


